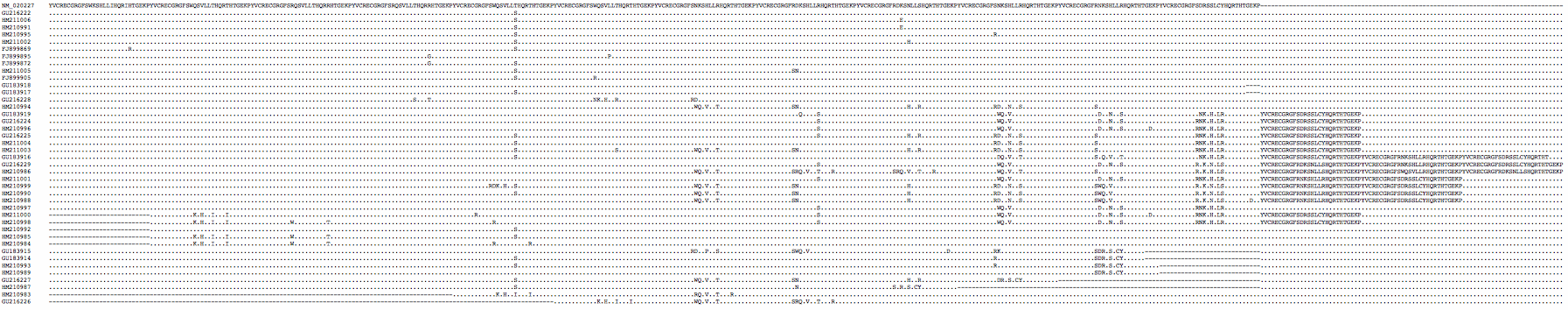PRDM9: meiosis and recombination: Difference between revisions
Tomemerald (talk | contribs) |
|||
| (261 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
'''See also:''' [[PRDM11:_giant_missing_exon|All about PRDM11]] | |||
'''Updates''': To help readers locate fixes, additions and other news as the article grows longer, significant | |||
additions will be noted here in reverse chronological order linked into their spot in the article. | |||
04 Feb 11: improved cow and sheep assemblies have [[#Curated_reference_sequences|six PRDM7/9 genes and a functional chrX copy with 21 zinc fingers]]. | |||
15 Dec 11: wake up folks: that gene in dog and mice is '''PRDM7''' [[#Correct gene tree for PRDM7 and its spin-off PRDM9s|not PRDM9]] | |||
11 Dec 11: added a whole [[PRDM11:_giant_missing_exon|new page on PRDM11]] -- the closest match in non-mammalian amniotes to the PR(SET) methylation domain of PRDM7/9 | |||
30 Oct 11: added 6 new fragmentary Carnivora PRDM7 sequences that -- like dog -- have inactive terminal exons. | |||
12 Sep 11: started [[#Origin_of_Species_and_all_that|new section]] on origin of species, sex chromosome co-evolution etc. | |||
11 Sep 11: re-edited first 20 pages for glitches, redundancy and inconsistencies. | |||
09 Sep 11: added sections on [[#Marsupials_and_platypus:_the_mystery_of_exon_5|chained meiosis]] in platypus and [[#Comparative_genomics_of_placental_mammals|non-PAR PRDM9 gene conversion]] sites on human chrY. | |||
07 Sep 11: transversional [[#Sequence_analysis_of_human_variation|second block]] of human PRDM9 recognizes hotspots. | |||
31 Aug 11: the first 523 amino acids of primate PRDM9 are not evolving at an [[#Rate_of_proximal_PRDM9_evolution_in_primates|anomalous rate]]. | |||
28 Aug 11: [[#Comparative_genomics_in_placental_mammals|partial distal pseudogenization]] of PRDM7 in some catarrhines. | |||
22 Aug 11: re-wrote section proving [[#Comparative_genomics:_sequence_availability|mouse PRDM9 is really PRDM7]]; re-analyzed [[#Comparative genomics in placental mammals|historic species barrier]] paper. | |||
22 Aug 11: improved [[#Introduction|mouse PAR region]] depiction and updated [[#Comparative genomics: sequence availability|expression data]] -- retinal transcripts suggest a multi-functional protein. | |||
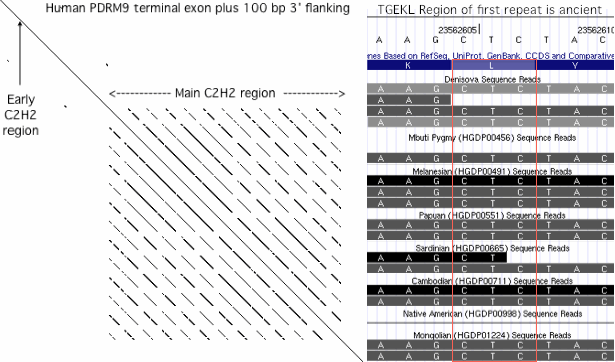
19 Aug 11: showed odd TGEKL cap region of first human repeat was already present in Denisova and pygmy. | |||
18 Aug 11: fixed timing of the primate PRDM7 duplication creating PRDM9 relative to marmoset, tarsier, lemurs and tree shrew. | |||
== Introduction == | == Introduction == | ||
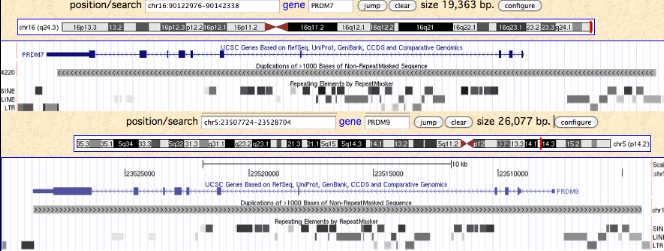
PRDM9 is a gene on human chromosome 5 with a very peculiar history. Its primary function -- after many false starts -- has only recently become clear: scanning the genome with its terminal zinc finger array to locate and mark recombination hotspots with its histone methylase where its transcription factor domain can direct additional proteins to initiate the double stranded breaks needed for meiosis. Some level of recombination between homologous chromosomes is essential to proper alignment and separation into daughter cells as well as for bringing favorable alleles onto the same haplotype for adaptive evolution. | PRDM9 is a gene on human chromosome 5 with a very peculiar history. Its primary function -- after many false starts -- has only recently become clear: scanning the genome with its terminal zinc finger array to locate and mark recombination hotspots with its histone methylase where its transcription factor binding domain can direct additional proteins to initiate the double stranded breaks needed for meiosis. Some level of recombination between homologous chromosomes is essential to proper alignment and separation into daughter cells as well as for bringing favorable alleles onto the same haplotype for adaptive evolution. | ||
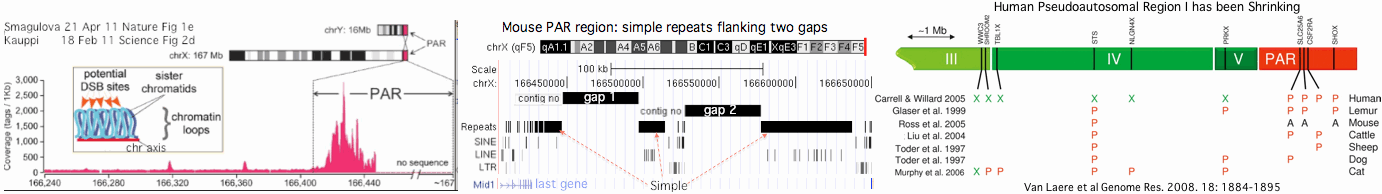
This reaches criticality in placental mammal sex chromosomes which are limited in homologous alignability to short pseudoautosomal regions (PAR). Here in male meiosis, a recognizable sequence site must be found for the double stranded break with only tens of kilobases available in mouse, the most favorable [http://www.ncbi.nlm.nih.gov/pubmed/21460839,21330546 experimental situation]. However two large gaps remain in the most recent mouse assembly used (July 07) telomeric to the single known PAR hotspot (a situation not improved in the [http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=10090 July 2011 release 37.2]) nor fixed by [http://www.pnas.org/content/108/4/1513.full Illumina reads]. This region likely consists entirely of sequence categorized as simple repeats, meaning that as one hotspot is erased by gene conversion, similar ones remain available in the region, mitigating the need for adaptation in PRDM7/9. | |||
[[Image:PrdmPAR.gif]] | |||
Humans are [http://genome.cshlp.org/content/18/12/1884.full unique] in having a second PAR of size 330 kb on distal Xq which contains five genes, [http://genome.cshlp.org/content/13/2/281.full acquired recently] by chrY via LINE-mediated illegitimate recombination. Shrinkage of the larger PAR1 (currently 2.7 Mb, 24 genes) since lemur divergence (a process at much longer time scales driven by [http://gbe.oxfordjournals.org/content/1/56.full strata-creating inversions]), reduces the potential number of recombination initiation sites and may correlate with autosomal duplication of PRDM7 around this same time. | |||
Gene conversion also occurs unexpectedly in human at various [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2706966/ hotspots] between the PARs, notably in the PRKY, [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2833382/ VCX/Y], TGIF2LX/Y and IR1 and P1 regions -- as well as [https://lra.le.ac.uk/bitstream/2381/9310/1/2011bowdengrphd.pdf intra-chromosomally] in the palindromic section of chrY. The PRKY site contains a [http://www.ncbi.nlm.nih.gov/pubmed/19165926 canonical PRDM9 recognition site], CCCCCCCTTCCCTC. If recombination intermediates resolve as translocation rather than gene conversion, infertile [http://www.omim.org/entry/278850 46,XX males] and [http://www.omim.org/entry/233420?search=233420&highlight=233420 46,XY females] result. | |||
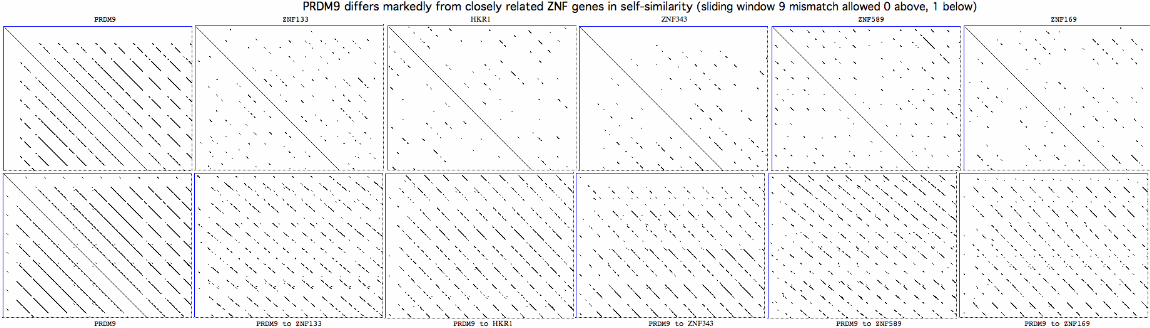
A protein central to an ancient essential process is usually highly conserved. However this is not the case here at all. Indeed, it proves exceedingly difficult to find a comprehensive set of PRDM9 orthologs even in the 39 sequenced placental mammalian genomes available on 10 Sept 2011, with immense and continuing confusion in the literature caused by independent segmental gene duplications, partial and full pseudogenizations and mix-ups with other composite domain proteins -- all compounded by outright sequencing error in the long terminal zinc finger repeat array. | |||
Although meoitic recombination is a universal feature of eukaryotes, surprisingly the underlying mechanism for it is not. As shown below, the PRDM9 scenario is not directly applicable even to other major placental mammal clades. And neither PRDM9 nor its parent gene PRDM7 have a full-length orthologous counterpart in monotremes, birds, lizards, amphibians or earlier diverging vertebrates. While similar domain combinations have arisen before in other narrow bilateran clades, no evidence connects them to meiosis. Drosophila in particular uses very different gene products; meiosis there does not involve zinc finger proteins. | |||
This puzzling history did not arise from ab initio sequence innovation in post-Cambrian deuterostomes because PRDM7 (the parent of primate PRMD9) arose instead from chimerization events in the mammalian stem involving SSX1, PRDM11 and a ZNF that together provided the six structural domains of the current protein. The parental gene histories are themselves complex, involving still earlier gene duplications and internal tandem repeat expansions -- patterns with numerous precedents in the overall metazoan proteome evolutionary context. Zinc finger proteins are a [http://www.ncbi.nlm.nih.gov/pubmed/21738707 greatly expanded, often chimeric family] within the mammalian lineage whose history may never be fully unravelled. | |||
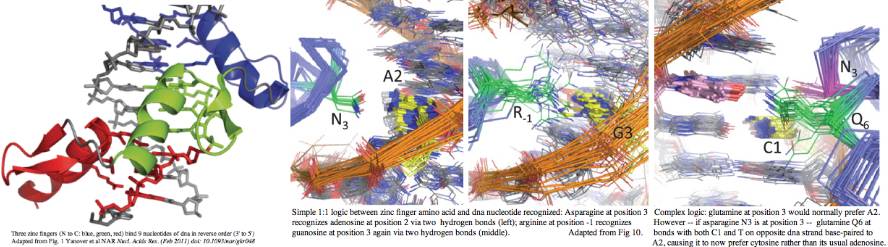
Rapid evolution of the terminal region of PRDM7/9 occurs at the amino acid level, both in the number of zinc fingers and -- within a given finger -- in the four non-adjacent residues primarily responsible for recognizing a specific dna trinucleotide. This variability is not coincidental to the role in meiosis -- that process tends to destroy its recombination hotspots by biased gene conversion. Since recombination is essential, new hotspots must emerge. The race is then on for PRDM7 or its spun-off PRDM9s to rapidly evolve and define new histone markup sites. | |||
The consequent mutational pattern is quite distinct from those of closely related zinc finger proteins. It may result from error-prone replication slippage in the repeat region but why that mechanism would concentrate non-synonomous change at certain residue positions remains '''unexplained''', along with [http://www.pnas.org/content/108/30/12378.long fine structure details] in meiotic data, transcription in non-meiotic retina and the functions of the conserved zinc knuckle and early zinc finger domains. | |||
This rapid evolution can cause breeding incompatibility between populations in the F1 generation (meiosis arrest for lack of cross-overs, notably between chrX and chrY) and thus be central to the process of speciation. However PRDM7/9 cannot provide a universal explanation of either speciation or Haldane's rule even within placentals because the hotspot-defining genes are not in straightforward correspondence. | |||
In effect, initiation of meiotic recombination is rapidly evolving -- and taking speciation and Haldane's rule along with it: each major clade of placentals has evolved a different hotspot recognition system, taking its most extreme form in pecoran ruminants with six PRDM7/9 genes (whose individual roles are not understood but could be quickly worked out in cattle). These differences within placentals follows upon the very different structure and gene content of sex chromosomes between monotremes, marsupials and placentals which in turn are quite different from those of birds, lizards and the amniote ancestor. | |||
Syntenic relationships can help resolve gene duplication events during mammalian evolution. Here the chromosomal gene order TUBB3+ AFG3L1+ GAS8+ has stably existed since the stem amniote some 310 million years ago, with PRDM7- qTer added in placental mammals after marsupial divergence and maintained there since over billions of years of observable branch length time. PRDM9 however is found in many syntenic contexts, depending on timing and positioning of the segmental duplications giving rise to these secondary copies. | |||
From the perspective of evolutionary genomics, PRDM7 is the fundamental gene, not the disparate collection of genes lumped under PRDM9 (even as those have taken over in primate meiois). At different times in different placental clades, PRDM7 spun off segmental duplications of itself to other sites in other chromosomes, probably because of a susceptible location at the extreme q arm of an autosomal chromosome. Because PRDM7 has stayed at its site adjacent to GAS8, it is possible to say unambiguously which of two initially identical copies was the parent gene. Due to this history, the 'PRDM9' genes do not form their own subtree within the overall two-gene tree under phylogenetic algorithms but instead associate more closely with their parental PRDM7 parent. | |||
These paralogous copies -- despite all being called PRDM9 -- are not usefully considered orthologous outside their species clade of origin. Orthology requires (by long-standing definition) vertical descent from a common gene '''in the last common ancestor''' of two species. Here primate PRDM9 are descended from a common gene (namely the recent duplicate of PRDM7 in the stem primate ancestor) but 'PRDM9' in afrotheres, pecoran ruminants, rabbits etc arose from different duplications at different times during placental mammal evolution from a rapidly evolving PRDM7 parental gene in those lineages and so -- despite the name -- are not vertically descended from a common primate PRDM9 in their last common ancestor (though all the genes here descend from a single stem placental PRDM7 gene). | |||
In tandem duplications, the parental copy cannot be distinguished but here the second copy was never on an equal syntenic footing. In a small segmental duplication, not all of the upstream regulatory features (which can be a megabase or more away from transcription start) are necessarily carried over. The second copy may be differently expressed from the get-go despite encoding an identical protein. | |||
In mammals, PRDM7 has segmentally duplicated numerous times, in each case to a different site on a different chromosome. While PRDM7 has a moderately long history, primate PRDM9 has none of its own prior to its creation in stem catarrhine. For clarity, the various PRDM7 duplications can be denoted PRDMPpri, PRDM9pec, and PRDM9afr. | |||
Such gene duplicates are [http://ai.stanford.edu/~serafim/CS374_2006/papers/Sonhammer_TIGs_2002.pdf sometimes called] in-paralogs within a species and co-orthologs across species. However these terms do not reflect the mechanism of duplication and are topologically unstable (depend on the species range included in the gene tree unlike the terms ortholog, paralog and homolog) and have gained limited traction. Synteny creates an asymmetry here because only the parental gene assuredly has all upstream and downstream effectors. | |||
Composite-domain proteins such as PRDM7/9 give rise to a whole new level of terminological muddle as each domain may have its own complex evolutionary history of earlier duplication, shuffling partners, and functional drift. It is very difficult to capture these histories within gene nomenclature. | |||
=== Comparative genomics of placental mammals === | |||
Within Euarchonta, a small segmental duplication encompassing PRDM7 took place in a stem catarrhine primate. The duplicated gene, designated PRDM9 by the human gene nomenclature committee, resides within an altogether new syntenic context -- a cadherin gene complex on an unrelated autosomal chromosome. PRDM9 initially shared meiotic functionality with PRDM7 even as it diverged in amino acid sequence and descended through speciation events into contemporary old world monkeys and great apes. PRDM7 still persists at its original ancestral location (qTer and adjacent to GAS8) but as an overt full-length pseudogene in some lineages (rhesus, gibbon) but not so clearly in others (orangutan). Gene duplication followed by lineage-specific reallocation of functionality is an exceedingly common scenario within metazoan evolution. The timing of gene duplication here means only catarrhine primates have a PRDM9 gene. | |||
Human PRDM7, despite its 3 frameshifts in exon 9 and 10, may still retain N-terminal functions (those not requiring dna recognition by the zinc finger array). PRDM7 is sometimes treated as a conventional gene with splice 'isoforms' despite an internal direct tandem repeat of 88 nucleotides in exon 9 of the human reference sequence that throws off the reading frame and subsequent splice donor to exon 10, which itself has a frameshift (GGGG to GGG) in the second of its three zinc fingers. This is not an anomaly of the reference genome as it is shared across the 1000 Genome Project. The C-terminal pseudogenization of human PRDM7 [http://main.genome-browser.bx.psu.edu/cgi-bin/hgTracks predated divergence] of denisovan, bushman and neanderthal and postdated analogous events in other primates. | |||
Translation into an incorrect reading frame cannot plausibly yield a stable fold, much less zinc fingers that recognize a nucleotide meiotic sequence. Nothing is known about the fate of in vivo transcripts or mature PRDM7 protein but the likeliest scenarios are nonsense-mediated decay or proteolytic trimming of unfolded C-terminal rubbish. Partial pseudogenization is an option for a protein like PRDM7 with multiple quasi-independent domains. Transcripts and alternative splices -- being artifact-rich processes in vertebrates -- do not provide a reliable guide to any stably folded mature protein that may ultimately be produced. | |||
Initially PRDM7 must have shared its role in meiosis with PRDM9 (the sequences and near-upstream regulatory regions being identical), but later PRDM9 took over this role entirely in most primate clades as only it retained the zinc finger array. PRDM7 either retained non-meiotic roles (implied by numerous transcripts in non-meiotic tissue) or acquired other functionality not involving the terminal zinc finger array (but in some species losing all function). | |||
Chimp and gorilla have also lost functionality in the last exon. However the mechanisms of loss -- stop codon in chimp vs exon 10 frameshifts in gorilla -- differ from human. Orangutan PRDM7 may still be functional whereas gibbon is riddled with early stop codons implying total loss. Since pseudogenization is a fairly rapid process, it could not have begun at the time of segmental duplication in stem catarrhine. Instead PRDM7 co-existed with PRDM9 for tens of millions of years, only in the last few million years losing distal functionality independently in various great ape lineages. | |||
Loss of terminal array function took place fairly late in each great ape lineage by independent mutational mechanisms rather than by a shared disabling mutation in a common ancestor. Residual function may or may not be the same in those great apes that have retained the proximal portion of the gene. Partial pseudogenization is structurally acceptable in chimeric domain proteins if domains fold independently and do not significantly interact. | |||
This scenario is strongly supported by alignment of manually curated primate PRDM7/9 sequences up to but not including the final array. If PRDM7 had been inactivated early after duplication, it would have accrued a large number of non-synonymous changes by now, changes oblivious to the conservation status of individual residues in the early domains. The alignment [[#Rate_of_proximal_PRDM9_evolution_in_primates|below]] shows preferential retention of conserved residues, as well as PRDM7 and PRDM9 clustering into distinct gene sub-trees as expected from the time of duplication. Gene conversion can keep duplicated genes synchronized for a time but that mechanism is not applicable over such a time span for non-tandem genes on unrelated chromosomes. | |||
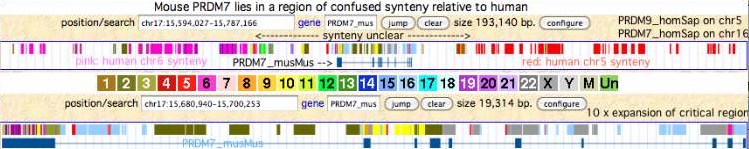
Earlier diverging primates such as new world monkeys and lemurs have a single PRMR7 gene adjacent to GAS8. Although a PRDM9 duplicate could occur within an genome assembly gap, a large multi-exon gap is implausible in multiple assemblies with respectable Sanger trace read coverage, especially given the pedestrian chromosomal location of catarrhine PRDM9. The tarsier situation however is syntenically unclear -- the gene occurs in five separate contigs, mostly single reads. Tree shrew also has unsatisfactory coverage (six exons spread out over two contigs and 3 unassembled traces, a string of Ns in the terminal zinc finger domain, and undeterminable synteny). These scattered contigs could conceivably represent two or more separate genes. | |||
[[Image:PRconfusedSyn.jpg|left]] | |||
<br clear=all> | |||
Within Rodentia, the single mouse PRDM7/9 gene lies in a region of confused synteny (relative to human) attributable to chromosomal rearrangements in the rodent clade. The browser screenshot above shows an unrelated region of human chr5 -- not the part that bears human PRDM9 -- as right-syntenic neighbor to mouse PRDM7. The left-syntenic human chr6 segment does not carry human PRDM7 (human chr16). The mouse gene thus has no informative neighbors (not even flanking debris) from GAS8 or cadherins. Similarly the mouse orthologs of GAS8 and cadherins do not contain PRDM7/9 debris. | |||
The | The rat gene occurs in the same syntenic context as mouse but other rodent genomes (including the new hamster assembly) are too incomplete for synteny to be assessed. Thus the genetic rearrangement taking PRDM7 from its location facing GAS8 to its current position in rodents cannot be accurately timed relative to rodent divergences. The rabbit assembly of Apr 2009 is still quite garbled in the PRDM7-related region and also contains a spurious assembly stutter duplication. The syntenic location is unlike mouse/rat or any other mammal and again there is no debris at the relevant locations in other species. The other lagomorph assembly (pika) is missing its first and last exon so provides no syntenic information. | ||
Thus the history of chromosomal rearrangements of PRDM7-like genes in Glires requires better assemblies in more species before gene rearrangements, gains and losses can be understood. It would be [http://www.pnas.org/content/108/4/1513.full more useful] to finish genomes already begun rather than generate thousands of additional fragmentary assemblies as in the 10k vertebrate genome project. | |||
Conceivably, ancestral euarchontoglire PRDM7 duplicated twice to its current locations in rodents and lagomorphs from the ancestral location adjacent to GAS8 with the parental gene later lost twice. However even in this non-parsimonious scenario, the mouse gene cannot legitimately be called PRDM9 because it is still not a strict ortholog of primate PRDM9. A simpler scenario envisions two lineage-specific chromosomal rearrangements of PRDM7 with no gene gain or loss, consistent with Laurasiathere outgroup data indicating a single copy of PRDM7 at euarchontoglire divergence. | |||
Since human PRDM9 arose in early primates as a gene duplication of a much older PRDM7, it was not present at the time of mouse/human divergence (Euarchontoglires). Hence the mouse gene cannot correspond to it. The mouse gene is best taken as a straightforward ortholog of primate PRDM7. Mouse has a great many chromosomal rearrangements relative to ancestral Euarchontoglires, so a translocation here of PRDM7 is unremarkable. The mouse gene is still called PRDM9 in the scientific literature despite the Jan 2002mouse assembly establishing its lack of synteny to the catarrhine-specific gene! | |||
The | The PRDM7 protein is evolving conservatively in murid rodents though rather rapidly in the amino acids contacting the hotspot dna motif. There are substantial differences between common strains of lab mouse and unsurprisingly these cannot always interbreed (shown below in first six lines as genome strain C57BL/6J, WSB/EiJ, MOLF/EiJ, PWD/PhJ, CAST/EiJ, and C57BL10.F). Note mouse strains vary considerably in the number of zinc finger repeats -- 11 in CAST/EiJ, 12 in the reference genome strain C57BL/6J, 13 in C3H/HeJ and 14 in strain PWD/Ph -- and so in their dna-contacting residues. | ||
The species barrier between B6 and C3H mouse strains is said [http://www.sciencemag.org/content/323/5912/373.full entirely attributable] to a single difference in the number of zinc finger repeats (loss of repeat 10 in B6). As the extra repeat in C3H intercalates a new set of dna-contacting residues, its array recognizes a dna sequence with a 3 bp insertion relative to the recognition sequence of B6 (ie the barrier does not arise from repeat number variation per se). However it has not been established why a mouse heterozygous for separately meiotically functional PRDM7 genes cannot carry out meiosis. | |||
( | |||
While the species barrier result implicates distal zinc fingers in meiotic recombination, no meiosis occurs in retina, the primary source of (unsought) mouse PRDM7 transcripts at GenBank. Mouse PRDM7 may be [http://en.wikipedia.org/wiki/Protein_moonlighting multi-functional], with a distinct role regulating gene expression in retina (in the manner of conventional ZNF genes). The proximal block of zinc fingers could be used there for dna recognition, giving rise to dual (or overlapping) selection on the array. | |||
While consistent with meiosis site recognition requiring less than half of the array, a conflict arises because the mysterious mechanism generating mutational variation specifically at the dna-contacting residues for meiosis would be maladaptive for continuing recognition of fixed retinal gene regulatory targets (if any). Here it is imperative to understand what other genes are acting upstream to confine PRDM7 expression to testis and retina. If transcription in retina is specific to one of its many structural components, that could provide insight into its functionality there. | |||
[[Image:MouseSpeciation.gif|left]] | |||
<br clear=all> | |||
PRDM7_musMus1 SIERQCGQYFSDKSNVNEHQKTHTGEKPYVCRECGRGFTQNSHLIQHQRTHTGEKPYVCRECGRGFTQKSDLIKHQRTHTGEKPYVCRECGRGFTQKSDLIKHQRTHTGEKPYVCRECGRGFTQKSVLIKHQRTHTGEKPYVCRECGRGFTQKSVLIKHQRTHTGEKPYVCRECGRGFTAKSVLIQHQRTHTGEKPYVCRECGRGFTAKSNLIQHQRTHTGEKPYVCRECGRGFTAKSVLIQHQRTHTGEKP-YVCRECGRGFTAKSVLIQHQRTHTGEKPYVCRECGRGFTQKSNLIKHQRTHTGEKPYVCRECGWGFTQKSDLIQHQRTHTREKP-------------------------------------------------------- | |||
PRDM7_musMus2 ....................................................................................................................................................................................................................................................................................................A..V..Q.................R......N..K......G...YVCRECGWGFTQKSDLIQHQRTHTREK............................. | |||
PRDM7_musMus3 ........................................K.D..K........................V....................................................A..N..Q.....................A.....Q.....................Q..D..K..............................................................................E..S................................................R...A..V.........G........................................................... | |||
PRDM7_musMus4 ........................................K.D..K........................V....................................................A..N..Q.....................A.....Q.....................Q..D..K.................................................E..S..K.........................N...........................V....................R...A..V.........G........................................................... | |||
PRDM7_musMus5 .......................................AK.N...........................V..Q.................................................A..N..Q.....................E..S....................W......N........................Q..S..K........................N.............A.......W...Q..N..K.................W......D..Q......R..-----------------------------........................................................ | |||
PRDM7_musMus6 ...............................................................................................V..V.........................N.H..Q.....................A.....Q.....................QN.H........................Q..D..K.....................Q.....K......................Q..N..K.................W......D..Q......R..-----------------------------........................................................ | |||
PRDM7_musMol2 .......................................AK.N...........................V..Q.................................................A..N..Q.....................E..S....................W......N........................Q..S..K........................N.....................W...Q..N..K.................W......D..Q......R..-----------------------------........................................................ | |||
PRDM9_musCas .......................................AK.N...........................V..Q.................................................AR.N..Q........................D...........................N........................E..S..K.................W......N.........................Q..S..K.....................A.....Q........................N..K......G...YVCRECGWGFTQKSDLIQHQRTHTREKP............................ | |||
PRDM9_musPah ....................R...................K.N..T.....................G..P..R........................N..T.....................G..P..R........................H........................E..N..K.....................Q..P..R.............T.......Q..N..T....N......------------------------------------------------------------------------------------........................................................ | |||
PRDM9_musMac ........................................K.D..K.....................V..............................N..Q........................D........................V..H.TQ.....................Q..D..K........................H..K.....................Q..N............................N..K......................N.H.TQ.........S..........K......................................................................... | |||
PRDM9_musSpi ........................................K.N............-..............N..Q.....................A...........................V..H.TQ........................D...........................H.T......................Q..............................N..K......................QN.H.T..........S.......W..K...D..Q......R...----------------------------........................................................ | |||
PRDM7_musMol1 .......................................AK.N...........................V..Q.................................................A..N..Q.....................E..S....................W......N........................Q..S..K........................N.....................W...Q..N..K.................W......D..Q......R...----------------------------........................................................ | |||
PRDM9_merUng GTG.E...C.......S...R.................M.R.N..S....................M.R.N..S.....................V..V..S.....................V.PH..S..........H...........R.N..R.....................V.PH..S.....................V.PH..S.....................V.....S......................V.....R................R.....R.T..R..........H......R...RG.H.LR......G.VL........................................................ | |||
PRDM9_micAgr RVGGER..C...........R..................RK.N.NV.....................R.AL..S.......................AL..S........................Y..L.....................G..N.NV.....................Q..Y..L.....................G..L..R.....................Q..YP.L...........------------------------------------------------------------------------------------........................................................ | |||
PRDM9_arvTer RV.GE...C.N.......R.R..................RK.V..L........................V..N........................H..F........................H..L.....................W.....L........R............R..H..L.....................Q..H..L.....................R.....L......................R.....N..........--------------------------------------------------------........................................................ | |||
PRDM9_perPol R..TE...R.........S.R..SE..........Q..I.K.V..C.................Q...W..H..R.................K..IR..H..C.................Q..I...H..C.................Q.........C.................Q..IR..Y..C.................K...W..V..R......V...----------------------------.------------------------------------------------------------------------------------........................................................ | |||
PRDM9_perLeu R..TE...R......A..S.R..SE..........Q...RK.Y..C.................Q..I...V..R.................Q...R..Y..C.................Q..I......R.................Q...W.....C.................Q...R..Y..C.................Q...W..H..R.................Q...R..Y..C..................Q..IQ..H..C.................Q...R..Y..C.................Q...W..V..R......A........................................................... | |||
PRDM9_perMan RT.TE...H......A..S.R..SE..........Q...WK.V..R.................Q...W..V..C.................Q...W..V..C.................Q..I...H..R.................Q..IR..H..C.................Q..AQ.....Y.................Q...R..H..C.................Q..AQ.....C..................Q...W.....C.................Q..I...H..R.................Q..I...H..R......G........................................................... | |||
PRDM9_apoSyl RV...R..C.......S.R.G.......C...........K...NR..........H.............H.NR..........H..........L..N.NR..........C.......A.....D..Q........................N.NQ.....................R..L........................Q..D.NR...................L.Q..N.........................L..D........................R..D.NR.................R......N.........G...YVCRECGRGFTLKSDLIQHQRTHTGEKPYVCRECGRGFTRKSDLNRHQRTHTGEKP | |||
PRDM7_ratNor R.......C.......S...R........I........S.K.D..K......E....I......................E....I...........................I............D.........E....I............S..R...........I.....L...Q..N..R.L.........I.....L...R.................I.....Q.L.W..S...............I.........W..S.........V...--------------------------------------------------------........................................................ | |||
Laurasiatheres have a quite different history of gene duplication. Many clades simply retain the ancestral condition of a single PRDM7 gene adjacent to GAS8. Vampire bat (but not brown bat) has an additional segmental duplication to a novel location that is today a pseudogene. Insectivores, perissodactyls and early-diverging artiodactyls (alpaca, pig, dolphin) have a single PRDM7 gene in ancestral syntenic location (when determinable), though some genes have too few zinc fingers to define genome-specific hotspots in the manner of primates. | |||
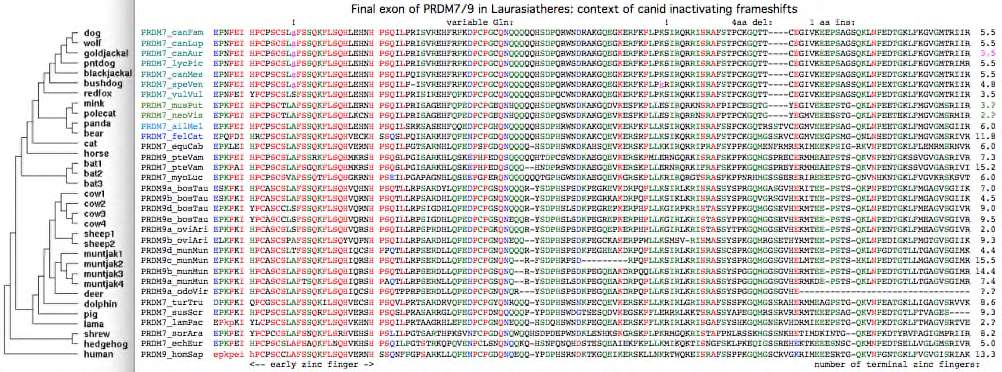
The dog reference genome inexplicably has a PRDM7 full-length pseudogene yet no PRDM9-like gene duplication despite a rather complete assembly, and possibly stable recombination hotspots according to a new [http://genome.cshlp.org/cgi/pmidlookup?view=long&pmid=22006216 low resolution study]. This is not an inbreeding artifact because five other species of canids share a frameshift within the early zinc finger of exon 7. Red fox (the outgroup) lacks this frameshift as well as a later shared frameshift in the third terminal zinc finger (thus timing them on the canid gene tree) but has an exon 7 frameshift of its own. | |||
Here it should be stressed that partial pseudogenization can only be ruled out in chimeric domain proteins by sequencing the entire gene, not done here. However based on several early inactivating mutations in dog, these canid PRDM7 are unlikely to be even partially functional. Thus some other mechanism must suffice for initiation of meiotic recombination in canids. Recall PRDM9 in humans explains only 40% of the events so a second mechanism (not necessarily that of canids) seem operative there too. | |||
PRDM7 from gray fox, the ultimate canid outgroup, unfortunately has not been sequenced and may still be functional. The alignment of exon 7 in 31 Laurasiatheres below shows rather few substitutions relative to the outgroup, establishing that pseudogenization is relatively recent. Thus PRDM7 was likely lost in canids around 8 myr ago and so was presumably functional during the 40 myr since divergence. | |||
However it is not so clear that PRDM7 is functional (in the sense of marking up meiotic recombination sites with a terminal zinc finger array) in the next outgroup (mink + bear). The first two repeats in the three species available have lost key conserved repeat residues for zinc binding and may be dysfunctional. Panda has an additional 3 repeats but this is still not sufficient to provide a recognition system of sufficient specificity as seen in mouse and human (or for that matter cat or bat). The ends of the mink and ferret PRDM7 genes are not satisfactorily covered by contigs so the number of zinc fingers there cannot be determined. | |||
Carnivores -- but not bats or horses -- have an intervening cadherin gene (CAD1) between GAS8 and PRDM7 which should not be confused with the weakly related cadherins (CAD10 and CAD12: 36% identity to CAD1) flanking primate PRDM9. This rare genomic event does not represent the ancestral state but is unfortunately too restricted in distribution to resolve the status of [[Pegasoferae%3F|Pegasoferae]]. | |||
Given the well-established phylogeny of [http://www.nature.com/nature/journal/v438/n7069/full/nature04338.html canids] within [http://wsbs-msu.ru/res/DOC185/carnivora.pdf carnivores], the first indel in the alignment below (a deletion of 4 amino acids) is perplexing because it unifies mink with canids which do not share a common ancestor to the exclusion of other carnivores according to the species tree. The tree topology may be slightly wrong (despite overwhelming statistical support), with mink sistered to canids, not bears. An early mutation followed by lineage-sorting seems implausible as the heterozygous state would have had to persisted 6 myr until mink/bear divergence. Convergent evolution -- an independent deletion in mink of the same length at the same site (respectively an independent insertion in bear) -- seems equally implausible unless special predisposing local dna attributes exist. Sequencing error can be ruled out since two mink, bear and panda, and seven canids provide a consistency check. The second indel -- a single amino acid insertion (serine) in ancestral canid -- is fully consistent with the gene tree. Given that this portion of exon 7 seems under little selectional constrain, neither indel is likely to have functional significance. | |||
[[Image:CanidExon.jpg|left]] | |||
<br clear=all> | |||
Difference alignment of exon 7 relative to dog shows relatively few substitutions given the rapid overall rate of evolution of the inter-domain region: | |||
PRDM7_canFam EPNPEIHPCPSCSLAFSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTT----CEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR | |||
PRDM7_canLup ................................................................................................................................................ | |||
PRDM7_canAur ................................................................................................................................................ | |||
PRDM7_lycPic ...................................................................S..........................................................................M. | |||
PRDM7_canMes ....................................Q........................................................................................................... | |||
PRDM7_speVen ....................................-................................................R..................................S....................... | |||
PRDM7_vulVul ......Y.......S........................I....Q....................C........................K..................................................... | |||
PRDM7_neoVis ..K.........T.............KC...........AG...Q.E.....E..H.........S...K.....V..S....LE......N....P.......G....Y..M.E..S.T.-......E.E...M....S.M.. | |||
PRDM7_musPut ..K.........T..............R...........AG...Q.E.....E..H.........N...K....DV..S....LE.....KN....PI..E...G....Y....E....T.-......E.....M....S.... | |||
PRDM7_ailMel ..K................................S.K.AS...QQE.....H...........H....K.....V.......L.............S......RSSTV...M.E......-......E.....M....SG... | |||
PRDM7_felCat ..Q.D..R.................V.CK.S..S..Q..A.K..Q.EN....D...........HS...K..C..V...SR..L...K...............MGSSRV...M.E.G..M.-.N..S.......M....S..V. | |||
PRDM7_equCab ..KL.....................V.R........GT.A.N.LQ.E..S..D......-....HS.K.K.HS..V...S...L.K......P....Y.P...MENFRMR.R.ME.K..I.-R.V.........LEMR.S.NV. | |||
PRDM9_pteVam ..K...............R......MKRS....S..G..A.K.LQS.E.H.ED.S......T..CS...K.E...V...S..MLERNG..K......K.P...MGSPRE..RMMEA...TS-..V...N...SSV...AS..V. | |||
PRDM7_pteVam ..K.A............G.......MKRS....S..G..A.K.LQS.E.H.ED.S...--.N..RS...K.E...V...S...LERN...K.F....K.P...MGSPREY.RMMEA...TS-..V...N...SSV...AS..VI | |||
PRDM7_myoLuc ..K..........V.....T.....GKR....E...GAPAGN.LQSEE.G.ER.......QTG.HG...K.E...V.G.S...L.R....GT...SFK.PNRHMGSSSER.R.RE....T.-.NV.HKN.....V..KRSKSVT | |||
PRDM9a_bosTau .SK.K....A...............VQ......T.L.P.A.DYLQ.E.....S.....R-Y...HSPS.KPE.R.V.D.PQ..L....LK.....S.YSPR..MGASGVH.R.TE.-..TS-..P.........M.A.VSG..K | |||
PRDM9b_bosTau .SK.K....A...............VQR.....T.L.P.A.D.LQ.E.....N.....R-Y...HSPS.KPE.RKA.D.PQ..L...KLK.....S.YSPR..VGRSGVH.R.TE.-..TS-............M.A.VSG..K | |||
PRDM9d_bosTau ..K.K.Y..A..C.S..........VQR.......L.P.IGD.LQ.E.....S.....R-Y...HSLS.KPE.R.P...PH..L.G..PK...T.S.Y.P...MGGSEVH.RMTE.-..TS-............MEA.VSG.V. | |||
PRDM9e_bosTau ..K.K.Y..A..C.S..........VQR.......L.P.IGD.LQ.E.....S..E..R-Y...HSLS.KPE.R.P...PH..L.G..LK...T.S.Y.P...MGGSEVH.RMTE.-..TS-............MEA.VSG.V. | |||
PRDM9a_oviAri ..K.K....A....S..........VQRS......L.P.P.D.LQ.E.....K.....R-Y...HSPS.KPE...P...PH..L.G..LK...T.S.YTP...MGGSEVH.KMTE.-..TS-......N.....MEA.VSG.V. | |||
PRDM9b_oviAri .LK.K....A...P..........YVQP.......L.P.A.D.LQ.E.....N..E..--Y...HSPS.KPE.CKA...PPW.L..MSV-...M.S.YSP...MRGSETHYRMTE.-..TS-.......I....M.T.VSG..K | |||
PRDM9d_munMun ..K.K....A....T..........IQCS..P.T.L.P.E.DLLQ.E.....N.....R-Y...HSPS.KPE.H.A.D.PQ..L....LK.....S.CSPR..MGGSGVH.RMTE.-..TS-.....G...T.LT.A.VSG.MK | |||
PRDM9c_munMun ..K.K....A...............IQRS....T.L.P.E.DLLQ.E.....N...--R-F...H.PS.---------.PQ..L....LK.....S.YSPR..MGGSGVH.LMTE.-..TS-H........T.LM.A.VSG.M. | |||
PRDM9b_munMun ..K.K....A...............IQRS....T.L.P.E.DLLQ.E.....S...--R-Y...HSPS.KPE...A.D.PQQ.L....LK.....S.YSPG..MGGSGVH.RMTE.-..TS-.........T.LT.A.VSG.M. | |||
PRDM9a_munMun ..K.K....A......T........IQRS..A.T.L.P.E.NLLQ.EH....S...--R-Y...HSLS.KPE...A.D.PQ..L....LK.....S.YSPG..MGGSGVH.RMKD.-..TS-.........T.LT.A.VSG.M. | |||
PRDM9a_odoVir ..K.K....A...............IQCS....TP..P.E.DLLQ.E.....N...R---Y...HSPS.KPE...A.D.PQ..L....LK.....S.YSPG..MGGSGVH.--------------------------------- | |||
PRDM7_turTru D.K.K.Q..G..........I....V.CS....V...T.A.DRVQ.E.....Y..R...-Y...HS.SNKPEC..V...S...L.R..LG.......SSP...MGSSRAH.RMMEAG..T.-..V...A....LI.A.VS.VVK | |||
PRDM7_susScr ..K.K..............R.....V.RS....S...A.A.RGLQ.EG...DN.Q...P-YP..HS.DGTSES.DV..GS..FLERR.L.KT...S.YAPE..MRSSRVR.RMTE......-..V......T..TVA.ES---- | |||
PRDM7_lamPac --E.K.YL.................VK..........TAAGR.LE.E.....N..E...-...QHS...KPE...A...S..FL.R..L....G...YSH...MGNSRVHDRMIE....T.-..V..K......TWA.VS.TVE | |||
PRDM7_sorAra ..K...Y...C......N.....R.V..S...L...GT.A.T.PKSVNF...D...W..HSDPDEP...KLENHKS.G.S.....RMG.K...T..PNLRSSKMGSSNKH.T.MDKINTG--..E..K..YRV.A.I..P.... | |||
PRDM7_echEur ..K..............A....N..VK.S.......GT.T.KQPQVEN..LSN....K.-..NF.NQH.STES..AI.K....L.M.K.KT..NG..KLP.E.IGSSREH.KTKE..-.NSC..M.....SE.LV.L..S..V- | |||
PRDM9_homSap ..K.........C............V.R..S..NF.GP.A.KLLQ.EN....D...E..-YP..HSR..KT....I...S.L.N.RTW..E......S.P...MGSCRVGKR.ME..SRT.-..V..GN.....V...IS..AK | |||
PRDM7_canFam VKYRGCGRGFNDRSHLSRHQRTHTGE<font color=blue>N</font>P YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECG<font color=magenta>r</font>GFIQRSNLSIHQRTHTGEKP | |||
PRDM7_canLup VKYRGCGRGFNDRSHLSRHQRTHTGE<font color=blue>N</font>P YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECG<font color=magenta>r</font>GFIQRSNLSIHQRTHTGEKP | |||
PRDM7_canAur VKYRGCGRGFNDRSHLSRHQRTHTGE<font color=blue>N</font>P YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECGRGFTQRSTLNEHQRTHTEEKP | |||
PRDM7_lycPic VKYRGCGRGFNDRSHLSRHQRTHTGE<font color=blue>N</font>P YVCRECGRGFTHRTNLIIHQRTHTGEKP YVCRECG<font color=magenta>r</font>GFIQRSNLSIHQRTHTGEKP | |||
PRDM7_canMes VKYRGCGRGFNDRSHLSRHQRTHTGE<font color=blue>N</font>P YVCRECGRDFTHRTNLIIHQRTHTGEKP YVCRECG<font color=magenta>r</font>GFIQRSNLSIHQRTHTGEKP | |||
PRDM7_speVen VKYRGCGRGFNDRSHLSRHQRTHTGE<font color=blue>N</font>P YVCREC<font color=magenta>g</font>RGFTHRTNLIIHQTTHTGEKP YVCRECG<font color=magenta>r</font>GFIQRSNLSIHQRTHTGEKP | |||
PRDM7_vulVul VKYRGCGRGFNDRSHLSRHQRTHMGE<font color=blue>N</font>P YVCRECGRGFTHRTNLIIHQRTHTGEKP YVSWECGRSFTRRSNLITHQRTHTGEKP | |||
PRDM7_neoVis VKYRG<font color=red>S</font>GQGFDDRSHLSRHQRTHKEEKP <font color=blue>S</font>V<font color=red>G</font>KE<font color=red>L</font>RREFIHKSVLVTHQRTHT<font color=red>E</font>ALP | |||
PRDM7_musPut VKYRG<font color=red>S</font>GQGFDDRSHLSRHQRTHKEEKP <font color=blue>S</font>V<font color=red>G</font>KE<font color=red>P</font>RREFIHKSVLVTHQRTHTGEKP YVCRECGRGFTQRSHLIRHQR | |||
PRDM7_ailMel VKYRGCGR<font color=red>D</font>FSDRSHQSGHQRRH<font color=red>-Q</font>KKP <font color=blue>S</font>VCKK<font color=red>V</font>KREFSHKSVLITHQRTHTGEKP YVCRECGRGFTQRSNLIRHQRTHTGEKP | |||
PRDM7_felCat IKNRGC<font color=red>E</font>QGFNDRSHFSRHQRTHKEEKP <font color=blue>S</font>VCNE<font color=red>F</font>RRDFSHKSALITHQRTHTGEKP YVCRECGRGFTQRSNLFRHQRTHTGEKP | |||
PRDM7_equCab VQYGGCGRGFNDRASLIKHQRTHTGEKP YVCRECEQGFTQKSSLIAHQRTHTGEKP YVCREC<font color=red>E</font>QGFSEKSHLIRHQRTHTGEKP | |||
PRDM7_pteVam VKYGGC<font color=red>E</font>HGFDDGSHLIMHQRTHSGEKP FVCRECERGFSKKSNLITHQRTHSGEKP FVCREC<font color=red>E</font>RGFTRKSSLITHQRTHSGEKP | |||
PRDM9_pteVam VKYGGCGHGFDDGSHFIRHQRTHSGEKP FVCRECERGFNEKSSLTMHQRTHSGEKP FVCREC<font color=red>E-</font>GFSVKSSLIRHQRTYSGEKP | |||
PRDM7_myoLuc IKHGGCGQGFNDGSHIDTHQRTHSGEKP YICRECGGFTHKSDL IRHQRTHSQENP YVCRECGRGFRDRSTLITHQRTHSGEKP | |||
gene_genSpp %id chr strand start stop span | |||
<font color=blue>PRDM7_canFam</font> 82% chr5 ++ 66560684 66567275 6592 <font color=blue>dog</font> | |||
<font color=blue>CAD1</font> 75% chr5 ++ 66571832 66581008 9177 | |||
<font color=blue>GAS8</font> 93% chr5 +- 66587321 66604940 17620 | |||
<font color=blue>PRDM7_ailMel</font> 100% GL193502 +- 628987 644235 15249 <font color=blue>panda</font> | |||
<font color=blue>CAD1</font> 73% GL193502 +- 620344 624223 3880 | |||
<font color=blue>GAS8</font> 91% GL193502 ++ 594843 609901 15059 | |||
<font color=blue>PRDM7_felCat</font> 100% Un_ACBE01450414 +- 10493 13105 2613 <font color=blue>cat</font> | |||
<font color=blue>CAD1</font> 75% Un_ACBE01450414 +- 3902 4280 379 | |||
<font color=blue>GAS8</font> | |||
<font color=red>PRDM7_equCab</font> 100% chr3 +- 36378853 36387224 8372 <font color=red>horse</font> | |||
<font color=red>GAS8</font> 93% chr3 ++ 36348528 36361906 13379 | |||
<font color=red>PRDM7_pteVam</font> 100% ABRP01250178 +- <font color=red>bat</font> | |||
<font color=red>GAS8</font> ABRP01250178 ++ | |||
<font color=red>PRDM7_myoLuc</font> 100% AAPE02062260 +- <font color=red>bat</font> | |||
<font color=red>GAS8</font> AAPE02062260 ++ | |||
Pecoran ruminants (cow, sheep, muntjak) present a vastly more complicated situation. Cows -- even in the revised assembly -- have a PRDM7 pseudogene adjacent to GAS8 accompanied by 5 PRDM9 copies in other locations (all distinct from the primate cadherin secondary site). This is neither a recent development nor an artifact of domestication because a similar expansion is seen in provisional assemblies of sheep and muntjak (wild deer) but not dolphin, pig or vicuna, dating the expansion to stem pecoran ruminant within artiodactyls. It is not clear which if any (or several acting in tandem) of these gene copies play a role in recombination -- the primate paradigm for meiotic markup is not immediately applicable to these species. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/17322288 Atlantogenata] (Afrotheres + Xenarthra) have yet another history. Elephant (best of the five available assemblies) has three loci: an old PRDM7 pseudogene in GAS8 syntenic position, a seemingly functional PRDM9a with 12 terminal zinc fingers and novel syntenic location, and a fairly recent pseudogene PRDM9b. Extinct mammoth shows the same three genes with the same pseudogenization pattern. Although the sequences diverged separately after speciation, three identical inactivating mutations occur in both mammoth and elephant but not hyrax, thus dating gene loss within afrothere speciation. This is shown for exon 9 below: | |||
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGR 1 PRDM9_conSeq wildtype consensus reference | |||
<font color = green>1 YVNCIQD*KEQNLVAFQYHRQIFHWTCCTIRPGCELLVWYGDNYSQELGIKWGSR<font color = red>*</font>KKELTSGT 1 PRDM9b_loxAfr gg bad acceptor, early stop codon, internal stop codon</font> | |||
<font color = green>1 YVNCTRDKEEQNLVAFQYHRQIFYWTCHTIQPGCelLVWYGDNYGQELGIKWGSR<font color = red>*</font>KKELTSGT 1 PRDM9b_mamPri gg bad acceptor, two 1 bp deletions, internal stop codon</font> | |||
<font color = green>1 YVRRARDTEERNLVAFQYHRQIFYRTCCTVRPGCELLVWRGAEDSQALG SRRTMELTSQK 1 PRDM9b_proCap pseudogene with 4aa deletion</font> | |||
<font color = brown>1 YVNCARDEEEQNLVAFQYHRQIFYRTCRTIQPDCELLVWYGDEYGQELGIKWGSRWKKELTSGT 1 PRDM9a_loxAfr wildtype</font> | |||
<font color = brown>1 YVNCARDEEEQNLVAFQYHRQIFYRT 1 PRDM9a_mamPri fragmentary coverage</font> | |||
<font color = brown>1 YVNCARDEDEQNLVAFQYHGQIFYRTCRPVQPGCELLVWYGDEYGQELGIQRGSRQMKALSSQT 1 PRDM9a_proCap 17 zinc fingers</font> | |||
<font color = blue>1 YVNGTQDEKEQNLVFFQYHRQIFYQTCYAVWPGCQLLVWYRDECGQELGIKWDNRGKKEFTVGT 1 PRDM7_loxAfr bad acceptor, bad donor</font> | |||
<font color = blue>1 YVNGTQDEKEQNLVFFQYHRQIFYQTCYAVWPGCQLLVWYRDECGQELGIKWDNRGKKEFTVGT 1 PRDM7_mamPri bad acceptor, bad donor, 1 synon bp difference</font> | |||
<font color = blue>1 YVNCARDDEEQNLVAFQYHRQIFYRTCRAIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAEK 1 PRDM7_choHof wildtype</font> | |||
<font color = blue>1 YVNCAWDDKEQNLVAFQYHRQIFYRTCRTIRPGCELLVWYGDEYGQELGIKWGSKWKKEFMTGT 1 PRDM7_dasNov wildtype</font> | |||
=== Correct gene tree for PRDM7 and its spin-off PRDM9s === | |||
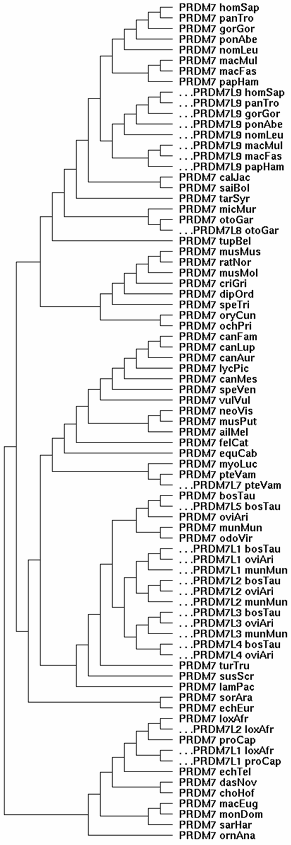
[[Image:PRDMLtree.gif|left]] | |||
At left is the gene tree summarizing the evolutionary relationships of the 80-odd PRDM7/PRDM9 homologs currently available from GenBank genome and targeted sequencing projects. Since the mammalian species tree is well-known, the gene tree can be clamped to it rather than derived ab initio (which won't work because some loci that must be included are pseudogenes in various states of degeneration. | |||
The gene tree shows PRDM7 (the fundamental gene here) spinning off various copies of itself at various times in various lineages over the last 102 million years of placental mammal evolution. Each spin-off has been offset and given a slightly different nomenclature to remind us that the primate PRDM9 gene duplication (called PRDM7L9 in the figure) should branch off at old world monkey and bear no special relationship to unrelated spin-offs in artiodactyls and afrotheres. The genes without offsets are conventional orthologs in the GAS8 PRDM7 qTer syntenic position, which has been stable for several billion years of summed branch length geologic time. | |||
These spin-offs had different subsequent histories. In some lineages, like primates, the spin-off PRDM9 took over from a pseudogenized parental gene PRDM7; in others, it became a pseudogene; in others, both genes seem to have persisted. | |||
Note neither carnivores nor rodents ever had a gene duplication of PRDM7. These species do not now contain a counterpart to human PRDM9 and never did. There is no evidence whatsoever for surviving pseudogene debris between CDH12-CDH10 (the syntenic location of all primate PRDM9), despite the extreme sensitivity possible with localized tBlastn searching. | |||
The | The fact that neither dog nor mouse has PRDM9 is not about naming conventions. It is about shifting functionality and shifting mechanisms for initiating meiotic recombination over evolutionary time. To discuss that intelligently, the genetic loci involved are best given names that are in themselves informative about gene evolutionary relationships, ie be compatible with their placement in the gene tree. That in a nutshell it is wrong to call the dog homolog PRDM9. In fact, it clusters by its synteny with the PRDM7s (including human) whereas the primate PRDM9s are off on their own subtree. (Put another way, if this is dog PRDM9, then where is dog PRDM7?) | ||
Human gene names are set by definition by HUGO and used exclusively in scientific journals by international agreement, the idea being to create a stable terminology (human genome is complete) and to avoid endless lab-specific synonyms, use of greek letters, roman numerals, subscripts, superscripts, upper/lower case, hyphens etc unsuitable for the bioinformatic era. | |||
HUGO attempts to give full length paralogs (above a certain percent identity) the same base name followed by consecutive numbering. This is not done consistenly as seen from PRDM* which is a pseudo-family of 16 genes with only a small PR(SET) domain in common (or PRNP/PRND which as full length homologs should have been PRNP1 and PRNP2). Pseudogenes are sporadically named. Despite the many flaws in HUGO names, it is unlikely that they will ever be significantly revised from where they are now. | |||
However the international agreement does not provide for official names or acronyms for the corresponding proteins and here the Wild West usage still prevails in journals. Just attaching a p as in PRDM9p to the HUGO name would work but people who work only at the protein level don't cooperate. However there is increasing pressure to standardize to allow computer mining of the biomedical literature. PubMed could require abstracts to contian a tag that would allow Blast searches against them (ie get around nomenclature variation). So far that hasn't happened. | |||
While human gene names are by definition correct, their underlying gene models are not. HUGO does not involve itself in gene curation but simply defers to a RefSeq at NCBI (which has no formal mechanism to correct errors). There are legitimate issues involving the significance, if any, of alternative splices and exon skipping and when something is a pseuogene. Thus human olfactory genes are difficult to sort out; in folate metabolism, a former DHRF pseudogene got upgraded to DHFL1 only in Oct 2011. | |||
All of the above just applies to humans, no other vertebrates. Mouse has its own official gene nomenclature committee (based at Jackson Laboratories). Here lower case is used for orthologs of HUGO gene names when possible (eg Prdm7) which doesn't scale to the other 5400 mammals. | |||
So how should genes be named in other vertebrates? Note humans have lost quite a few genes relative to other species (120 or more relative to last common ancestor with chicken, including whole subfamilies of opsins). So those genes cannot inherit names from HUGO human nomenclature unless it is ghosted forward to include lost genes. | |||
However in other mammals, simple 1:1 syntenic orthologs to human genes can be given the same name (eg dog or cat or panda PRDM7). This may covers 15,000 genes, a good start. For a segmental translocative duplication to a different chromosome like the PRDM7 -> PRDM9 case, PRDM7 is the parent gene and human PRDM9 the secondary derived feature. The syntentic gene in old world monkeys can safely be called PRDM9 as well. PRDM9 never existed in Carnivora and was not lost. Use of PRDM9 there would require a strange and different type of ghosted nomenclature than for lost genes. | |||
So what to call the extra copies of PRDM7 in elephant? Recall they have an old PRDM7 pseudogene in GAS8 syntenic position, a fairly recent retroprocessed pseudogene, and a seemingly functional copy with 12 fingers which has nothing to do with the human PRDM9 syntenic location. | |||
The name for this lineage-specific duplication in afrotheres should not confuse it with the unrelated lineage-specific duplication in primates (PRDM9) because these gene clades have no special relationship. Primate PRDM9 will branch out from the PRDM7 gene tree at primates, but the elephant copy from within afrothere PRDM7s . | |||
The nomenclature proposal here follows the HUGO template for the folate gene DHFR. That is, PRDM7 and numbered PRDM7L for Lineage-specific duplication. Here the primate PRDM9 would be PRDM7L9, elephant might be PRDM7L1, PRDM7L2, PRDM7L3, PRDM7L4 etc etc for the duplications in artiodactyls. | |||
The tree will need periodic revision as new mammalian genomes come in or as existing loci are re-interpreted. The Newick format that generates the tree is: | |||
(((((((((((((PRDM7_homSap,PRDM7_panTro),PRDM7_gorGor),PRDM7_ponAbe),PRDM7_nomLeu),((PRDM7_macMul,PRDM7_macFas),PRDM7_papHam)),(((((._._.PRDM7L9_homSap,._._.PRDM7L9_panTro),._._.PRDM7L9_gorGor),._._.PRDM7L9_ponAbe),._._.PRDM7L9_nomLeu),((._._.PRDM7L9_macMul,._._.PRDM7L9_macFas),._._.PRDM7L9_papHam))),(PRDM7_calJac,PRDM7_saiBol)),PRDM7_tarSyr),(PRDM7_micMur,(PRDM7_otoGar,._._.PRDM7L8_otoGar))),PRDM7_tupBel),((((((PRDM7_musMus,PRDM7_ratNor),PRDM7_musMol),PRDM7_criGri),PRDM7_dipOrd),PRDM7_speTri),(PRDM7_oryCun,PRDM7_ochPri))),((((((((((((PRDM7_canFam,PRDM7_canLup),PRDM7_canAur),PRDM7_lycPic),PRDM7_canMes),PRDM7_speVen),PRDM7_vulVul),((PRDM7_neoVis,PRDM7_musPut),PRDM7_ailMel)),PRDM7_felCat),PRDM7_equCab),(PRDM7_myoLuc,(PRDM7_pteVam,._._.PRDM7L7_pteVam))),(((((((PRDM7_bosTau,._._.PRDM7L5_bosTau),PRDM7_oviAri),(PRDM7_munMun,PRDM7_odoVir)),((((._._.PRDM7L1_bosTau,._._.PRDM7L1_oviAri),._._.PRDM7L1_munMun),((._._.PRDM7L2_bosTau,._._.PRDM7L2_oviAri),._._.PRDM7L2_munMun)),(((._._.PRDM7L3_bosTau,._._.PRDM7L3_oviAri),._._.PRDM7L3_munMun),(._._.PRDM7L4_bosTau,._._.PRDM7L4_oviAri)))),PRDM7_turTru),PRDM7_susScr),PRDM7_lamPac)),(PRDM7_sorAra,PRDM7_echEur))) | |||
,(((((((PRDM7_loxAfr,._._.PRDM7L2_loxAfr),PRDM7_proCap),(._._.PRDM7L1_loxAfr,._._.PRDM7L1_proCap)),PRDM7_echTel),(PRDM7_dasNov,PRDM7_choHof)),((PRDM7_macEug,PRDM7_monDom),PRDM7_sarHar)),PRDM7_ornAna)); | |||
<br clear=all> | |||
The genusSpecies acronyms are in alphabetic order below: | |||
ailMel Ailuropoda melanoleuca (panda) | |||
bosTau Bos taurus (cattle) | |||
calJac Callithrix jacchus (marmoset) | |||
canAur Canis aureus (golden jackal) | |||
canFam Canis familiaris (dog) | |||
canLup Canis lupus (gray wolf) | |||
canMes Canis mesomelas (black-backed jackal) | |||
choHof Choloepus hoffmanni (sloth) | |||
criGri Cricetulus griseus (hamster) | |||
dasNov Dasypus novemcinctus (armadillo) | |||
dipOrd Dipodomys ordii (kangaroo rat) | |||
echEur Erinaceus europaeus (hedgehog) | |||
echTel Echinops telfairi (tenrec) | |||
equCab Equus caballus (horse) | |||
felCat Felis catus (cat) | |||
gorGor Gorilla gorilla (gorilla) | |||
homSap Homo sapiens (human) | |||
lamPac Lama pacos (llama) | |||
lycPic Lycaon pictus (painted dog) | |||
macEug Macropus eugenii (wallaby) | |||
macFas Macaca fascicularis (crab-eating | |||
macFas Macaca fascicularis (crab-eating | |||
macMul Macaca mulatta (rhesus) | |||
micMur Microcebus murinus (lemur) | |||
monDom Monodelphis domestica (opossum) | |||
munMun Muntiacus muntjak (muntjac) | |||
musMol Mus molossinus (wild mouse) | |||
musMus Mus musculus (mouse) | |||
musPut Mustela putorius (ferret) | |||
myoLuc Myotis lucifugus (bat) | |||
neoVis Neovison vison (mink) | |||
nomLeu Nomascus leucogenys (gibbon) | |||
ochPri Ochotona princeps (pika) | |||
odoVir Odocoileus virginianus (deer) | |||
ornAna Ornithorhynchus anatinus (platypus) | |||
oryCun Oryctolagus cuniculus (rabbit) | |||
otoGar Otolemur garnettii (galago) | |||
oviAri Ovis aries (sheep) | |||
panTro Pan troglodytes (chimp) | |||
papHam Papio hamadryas (baboon) | |||
ponAbe Pongo abelii (Sumatran | |||
pteVam Pteropus vampyrus (bat) | |||
ratNor Rattus norvegicus (rat) | |||
saiBol Saimiri boliviensis (squirrel monkey) | |||
sarHar Sarcophilus harrisii (tasmanian devil) | |||
sorAra Sorex araneus (shrew) | |||
speTri Spermophilus tridecemlineatus (squirrel) | |||
speVen Speothos venaticus (bush dog) | |||
susScr Sus scrofa (pig) | |||
tarSyr Tarsius syrichta (tarsier) | |||
tupBel Tupaia belangeri (tree shrew) | |||
turTru Tursiops truncatus (dolphin) | |||
vulVul Vulpes vulpes (red fox) | |||
=== Marsupials and platypus: the mystery of exon 5 === | |||
Tracking PRDM7 back to marsupials and beyond is problematic. The three available marsupial assemblies are seriously incomplete, causing gene prediction issues as exons are spottily represented and spread over multiple small contigs which cannot be tiled up into full-length genes, much less yield syntenic information. Because PRDM7/9 contain domains found in many other chimeric proteins, isolated exons cannot always be assigned correctly to their parent gene. | |||
Further, some exons in PRDM7/9 have weak amino acid conservation and so fail to give definitive blast matches to placental queries, a problem exacerbated for short exons and decayed pseudogenes (opossum). No expression data exist to bridge uncertain regions, meaning missing diverged exons cannot be located. Because the domains here occur widely in other combinations in other proteins, a full length marsupial sequence is critical to testing whether the domain shuffle resulting in PRDM7 and PRDM9 was a placental innovation. | |||
The most favorable situation occurs in the Monodelphis domestica assembly. Although exons 1 and 5 are missing, eight of the ten expected exons are readily located in a single assembly region of length 33,449 bp containing a single internal gap (estimated at 270 bp). It is not surprising that exon 1 cannot be located because it contains no known Pfam domain or reason for fixed length and diverges rapidly in placentals. However locating exon 5 is important for distinguishing between two adjacent small genes evolving into a single fused gene only in the placental branch versus a full length gene already present in the last common ancestor. | |||
Unless exon 5 lies within the assembly gap, it should be locatable in the 25,548 bp separating exon 4 and exon 6 (of which 8,263 bp remains after application of [http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker RepeatMasker]). However blastx against a panel of 54 exon 5 sequences from placental mammals fails to give any suggestion of match in any species, despite plausibly adequate length (all placental exon 5 sequences have 52 amino acids). | |||
Gene prediction tools such as GenScan, NScan, Ensembl and Gnomon give unsatisfactory results: a few exons are correctly predicted but are otherwise embedded in time-wasting rubbish. The poor reliability of these tools does not justify GenBank clutter (eg XM_001369137) providing their predictions. The 46-species whole genome alignment at UCSC (starting with PRDM7/9 'ProteinFasta' link at the description page) is a better starting point. | |||
Here it should be noted that exon 5 has not diverged especially rapidly from the last common ancestor of placentals. Aligned to human, the full range of sequences has overall identity of 69%. Exon 5 has a number of invariant and semi-invariant residues, only possible over this time span if maintained by selective pressure. Thus it has some function even though it contains no known Pfam domains and has no crystallographic structure match. | |||
Because exon 4 has a splice donor of phase 0 and exon 6 a splice acceptor of phase 2, the putative exon 5 in marsupials must take the form 0 xxx...xxx 1 to conserve reading frame. This rules out non-use of exon 5 in marsupials (alternative splicing) followed by mutational decay to unrecognizability. In the scenario of two adjacent genes not yet fused, the distal region would need a new exon containing the initial methionine and phase 1 splice donor because no iMet occurs in the extended reading frame of exon 6. | |||
The opossum gene is peculiar in that 7 of the 8 exons available are quite conventional in sequence but the terminal zinc finger exon is completely broken up by frameshifts and stop codons and barely recognizable. It could not be used to initiate meiotic recombination, yet no substitute homolog is at hand. The other exons return only PRDM7/9 as significant matches when back-blasted against the human genome establishing that they have not been confused with the many hundreds of partial homologs with KRAB, SSXRD, PR (SET) or C2H2 domains. However human (and other placentals) could easily have lost even better blast matches since divergence from marsupials. Thus it remains unclear whether marsupials have a full length counterpart to placental PRDM7. | |||
The Sarcophilus harrisii assembly is missing the same two exons but has a conventional terminal exon with an intact zinc finger region of seven repeats (with two distal frameshifts however). Here exons 2 occurs in contig AFEY01202902 and exons 3-4 in AFEY01156721 with 1,436 bp left over to host exon 5; exons 6-10 are found in a third contig AFEY01386448 with 8,331 bp available upstream for exon 5. It can't be established that these contigs are actually adjacent in the genome. The six exons comparable between tasmanian devil and opossum are 82% identical to each other as proteins and 67% identical to those of human, not indicative of anomalous or especially rapid evolution in the context of entire proteome rates. | |||
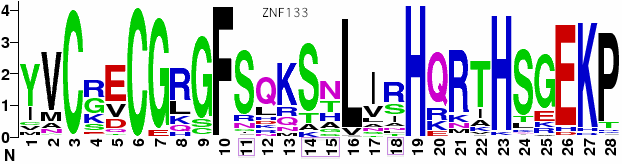
The Macropus eugenii (wallaby) assembly is least complete, with no contig containing more than a single exon. Here exons 1, 4, 5 and 8 are missing altogether but the terminal zinc finger exon is intact with 7 C2H2 domains. It is worth noting that the exon 10 is so long and distinctive with its phase 2 reading frame and early zinc finger that there is no possibility of confusing it with the closest human homologs (HKR1, ZNF133, ZNF169, ZNF343, ZNF589). However, humans could have lost an even better homolog of this exon. | |||
The gene adjacent to PRDM7 in mammals, GAS8, is an ideal probe, being single-copy and quite conserved in vertebrates. In the ancestral placental mammal, GAS8 and PRDM7 are convergently transcribed. Thus a marsupial contig containing the last exons of GAS8 might contain the last exon (or 3' UTR) of PRDM7. Even a partial exon or pseudogene remnant could be recognized with great sensitivity in such a contig. However none of the marsupial GAS8 contigs contain any information on PRDM7 or any other gene. | |||
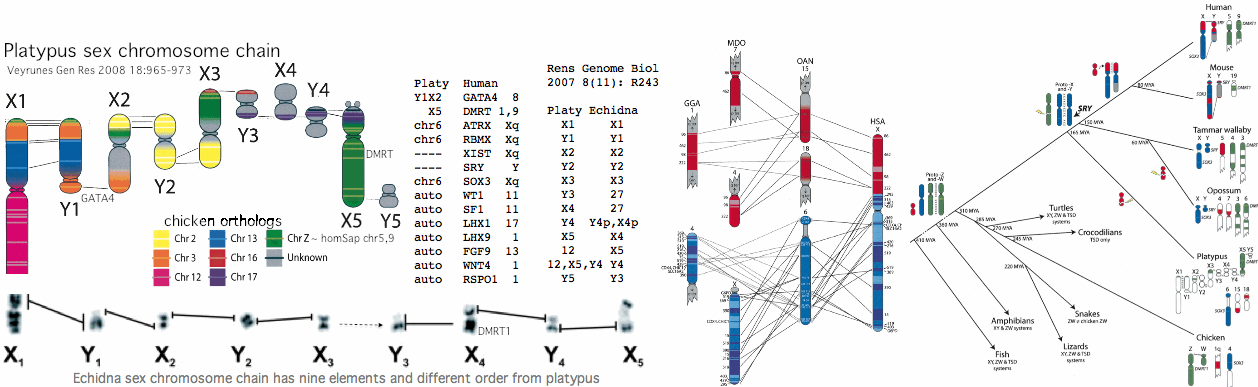
[[Image:PlatySxChr.gif]] | |||
The situation in platypus and echidna is curious according to [http://www.ncbi.nlm.nih.gov/pubmed/21250543,19874722,19874721,19874720,19874719,19874718,19802707,19196046,18983263,18606124,18463302,18185981,18021405,17717721,17400006,17317965,16344965,15723783 18 recent articles]. Note first that the chain of ten X and Y chromosomes (which is unprecedented in mammals) segregates during meiosis into either an X chain or Y chain, requiring crossovers in the 9 paired pseudoautosomal regions. Homology of key genes is to chicken, not theran mammals whose sex chromosomes thus arose after divergence at 165 million years. As with meiosis initiation, sex chromosomes seem never to stop evolving. | |||
It is not clear whether some form of PRDM7/9 is operative outside of placental mammal -- meiotic events have not been experimentally characterized to date in either marsupials or monotremes. Bird, alligator, and lizard (7 genomes) all lack candidate orthologs. Thus it is uncertain whether the massive restructuring of sex determination around this time correlates with the switchover to PRDM7/9 for meiotic recombination (in view of the sex chromosome recombination bottleneck) or is simply coincidental. | |||
In terms of platypus PRDM7/9 candidate orthologs, only distal exons 6-10 can be reliably recognized in the current assembly, ie KRAB, SSXRD and exon 5 are missing but the knuckle, PR and zinc finger domains are present with 3-4 repeat units. However the early zinc finger in the last exon is not present. Nonetheless, the best back-blast to human is still PRDM7/9. These exons occur in two tandem copies on the same strand but differ significantly from each other and so do not represent mis-assembly duplications. The intervening area is gapless so the missing exons should be locatable if present. | |||
< | |||
However they are not. Upon blastx of the repeatmasked sequence against Genbank tetrapod sequences, no matches occur, other than three worthless platypus gene models (XP_001507240, XP_001509482, XP_001509433) that predict earlier exons which however are wholly lacking in any support in any other species. Thus it appears that the gapless region does not contain any counterpart to exons 1-5 of theran mammals. Either this region has been lost in platypus or it is a stand-alone shorter distal version of PRDM7/9. | |||
< | |||
The first identifiable exons begins with the expected phase 2 reading frame in both tandem copies and do not contain an in-frame methionine upstream prior to a stop codon. Hence there must be at least one earlier exon. However tblastx of the appropriate regions of repeatmasked marsupial and platypus again does not identify noteworthy candidates. | |||
Perhaps the corresponding ancestral region was shuffled together with a gene providing the proximal regions in the theran branch only, giving rise to the full length gene there. However tblastn queries of the platypus assembly, while locating numerous appropriate KRAB_A domains with the correct 0 xxx...xxx 1 reading frame that back-blast to other human proteins, do not find counterparts of the exon 1-5 region beyond exon 2. Hence there is no obvious donor for the proximal half of PRDM7/9. | |||
Given that the PRDM and zinc finger families are greatly expanded with extensive domain shuffling in mammals with difficulties already tracing back PRDM7/9 to marsupials and monotremes, it comes as no surprise that bird, lizard and frog genomes shed no further light on the evolution of this gene. The situation in non-placental mammals could theoretically be resolved by sequencing transcripts, but these are exceedingly rare for PRDM7/9 even in placentals and so will not emerge unless explicitly sought. | |||
Conservation of exon 5 within placentals; invariant residues in <span style="color: #FF0000;">red</span> | |||
PRDM9_homSap GMPKASFSNE<span style="color: #FF0000;">S</span>S<span style="color: #FF0000;">LK</span>ELSRTANLLNASGSEQA<span style="color: #FF0000;">Q</span>KPVSPSGEASTSGQHSRL<span style="color: #FF0000;">K</span>L | |||
PRDM9_panTro .......N.........GMP....T............P.............. | |||
PRDM9_gorGor .....................................P.............. | |||
PRDM9_ponAbe .......N.........G.Q....T............P..........T..I | |||
PRDM9_nomLeu .................GA..................P.............. | |||
PRDM9_macMul .......N.......V.GM.....T............P...R.......... | |||
PRDM9_papHam E...T............G.P...ST.........A..P.............. | |||
PRDM7_calJac .......G......K..G...V..T..P.........P.............. | |||
PRDM7_micMur ...R.PL.DG.......G......T.....P......PR..........R.. | |||
PRDM7_otoGar ...R.PL.DG.......GP.S.P.I.....H..HM.SPR.........GR.S | |||
PRDM7_tarSyr ...R.PL.IV.......EM.....T.D....W......R.....E....K.. | |||
PRDM7_oryCun ...RLPVN.........GI.....TT...ED...SF.PK.TR......TR.. | |||
PRDM7_ratNor ET.RMPL.DK..V..VFGIE....T....H.....CSPE.GN.....FGK.. | |||
PRDM7_musMus ESSRMP..G..NV..G.GIE....T....HV.....SLE.GN......GK.. | |||
PRDM7_speTri LK.EVLL..........G......T.....V......LR...A.R....R.. | |||
PRDM9e_bosTau ..SR.PL.K.......PGA.K..KT..CK....L.P.PRK.R.PE..P.Q.V | |||
PRDM9c_oviAri ..S..LV..K.....MPGASK..KTR.PK...I..PAPR.P...E..P.Q.V | |||
PRDM9a_munMun ..SR.PLIK.......LGA.K.MKT...K...N..PHPRK.R.P...P.Q.V | |||
PRDM7_turTru AV.PVPL.......K.PGA.Q.QK...PA...S.AP.P.A....AW.T.Q.. | |||
PRDM7_lamPac ...RGPL..Q.......G..KP.KT...G.....FP.L.......R...Q.. | |||
PRDM7_susScr SDSRVPL..K......LT..EVPET.......E....P......RRR.GQE. | |||
PRDM7_canFam .I.RVPL..K.......E..K...T.SP..G..S..LP.K.....H.T.Q.. | |||
PRDM7_felCat .THRVPL.K.....DF.E..K...T.....G.....LP.......H...R.. | |||
PRDM7_ailMel .I.R.PLR.........E..K...T....LG.....LP.......HD.LQ.. | |||
PRDM7_musPut .V.R.PL..........E..K...T....HD.....HP.......H..LR.. | |||
PRDM7_pteVam A..RVPL...P......VI....K......D....F.P.K..A.R....Q.. | |||
PRDM7_myoLuc AKSR.PL..........G.....TT.....T..T.P.P.........P.S.. | |||
PRDM7_equCab R.RT.PL....R.....G..K..KT.S...V......L....S.E....R.. | |||
PRDM7_sorAra .RSRTPI.....S....G.RT...TKCTK.....LF.P.......HY.KP.. | |||
PRDM9a_loxAfr .T...LLG.......V.G..I...TT..........SP......D.P..W.. | |||
PRDM7_echTel ...GV.LR...N..V..G..I..T.AEP..PH-.G..P...T..HE.L.Q.V | |||
PRDM7a_proCap .T...LLG.......V.G..I...TT..........SP......D.P..W.. | |||
Consensus GMPRAPLSNESSLKELSGTANLLNTSGSEQAQKPVSPPGEASTSGQHSRQKL | |||
=== Comparative genomics | === Comparative genomics: sequence availability === | ||
[[ | As of Sept 2011, some 72 PRDM7 and PRDM9 genes from 45 species can be recovered from mammal genome projects. The encoded proteins are parsed into exons in the [[Curated reference sequences]] section. Due to gaps in coverage, full length gene models could not always be established. | ||
There has been no gain or loss of introns -- all genes have the same identically phased ten exons. No retroprocessed (intronless) genes occur in any species despite transcription in germline tissues. However 83 of 710 expected exons could not be found for lack of coverage or (for marsupials and monotremes) possibly too much divergence. | |||
In low coverage genomes, internal frameshifts and stop codons cannot easily be distinguished from sequence error even by consulting raw trace reads. However multiple disabling changes accompanied by an amino acid substitution pattern incongruent with conservation profile are strong evidence of pseudogenization, at least past the point of the first inactivating mutation. In chimeric proteins, the proximal region of the protein could continue to be functional. | |||
Since typically only one individual of unspecified gender of a species is sequenced on just one of two relevant autosomal chromosomes, an aberrant gene could reflect a bad heterozygous allele, an atypical homozygous individual (who might have impaired meiosis), or a balanced polymorphism that advantageously reduces copy number. A population survey is necessary to distinguish between these possibilities and overall SNP variation. In some genes, the zinc finger array seems too short to have sufficient site specificity. However these are known to contract and expand in the two intensively studied species (mouse, human), so here too the sequence from a single individual can be misleading. Without this data, it can difficult to say whether a given PRDM7/9 locus is a pseudogene. | |||
Supporting transcripts do not resolve the issue, first because pseudogene can continue being transcribed for millions of years after losing all functionality at the protein level and second because PRDM7 and PRDM9 are barely represented among the millions of mammalian transcripts at GenBank. That rarity might be explained by low levels of transcription in tissue types not widely used as experimental sources. However testis is frequently studied and one or more members of this gene family is essential for meiosis. This illustrates the futility of undirected transcript sequencing projects for determining the full coding potential of the genome. Global expression chips to date have not produced results here either. | |||
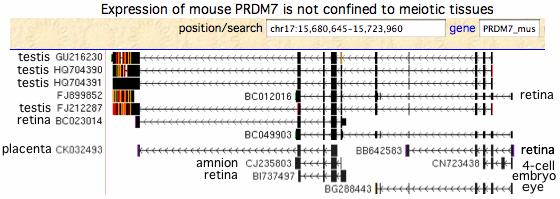
[[Image:MouseTranscripts.gif|left]] | |||
The transcripts from mouse, rat and pig do not support the widely propagated concept that PRDM7/9 function solely in meiosis (which would limit them in effect to testis) as most transcripts arise elsewhere. In mouse, the PRDM7 role in meiosis has strong experimental support, yet many transcripts come from non-meiotic tissues. Human PRDM9 experimental transcripts mostly derive from a single unpublished 2011 project entitled "Exhaustive RT-PCR and sequencing of all novel TWINSCAN predictions in human" which unhelpfully pooled tissue from adrenal gland, bone marrow, brain, cerebellum, brain (whole), fetal brain, fetal liver, heart, kidney, liver, lung, placenta, prostate, salivary gland, skeletal muscle, thymus, thyroid, trachea, uterus, and spinal cord with testis. | |||
<br clear=all> | |||
Transcripts at GenBank on 22 August 2011 (est database): | |||
<font color=blue>DB452778 PRDM9 homSap testis | |||
DB636359 PRDM9 homSap testis | |||
DB024448 PRDM9 homSap testis | |||
DB080053 PRDM9 homSap testis | |||
DT932634 PRDM9 homSap pooled including testis | |||
DT932633 PRDM9 homSap pooled including testis | |||
DV080525 PRDM9 homSap pooled including testis | |||
DV080526 PRDM9 homSap pooled including testis | |||
DV080328 PRDM9 homSap pooled including testis | |||
DV080173 PRDM9 homSap pooled including testis | |||
DV080174 PRDM9 homSap pooled including testis | |||
DV080327 PRDM9 homSap pooled including testis | |||
BU194881 PRDM9 homSap melanotic melanoma | |||
AL704902 PRDM9 homSap not reported</font> | |||
<font color=green>GU216230 PRDM7 musMus testis | |||
FJ212287 PRDM7 musMus testis | |||
HQ704390 PRDM7 musMus testis? | |||
HQ704391 PRDM7 musMus testis? | |||
CK032493 PRDM7 musMus placenta | |||
CJ235803 PRDM7 musMus amnion | |||
CN723438 PRDM7 musMus 4-cell embryo | |||
BI737497 PRDM7 musMus retina | |||
BB642583 PRDM7 musMus retina | |||
BC012016 PRDM7 musMus retina | |||
BC023014 PRDM7 musMus retina | |||
BG288443 PRDM7 musMus eye</font> | |||
FM103467 PRDM7 ratNor body fat | |||
GO353654 PRDM7a bosTau 4-cell embryo | |||
EF432551 PRDM7 bosGru testis | |||
BX673635 PRDM7 susScr pooled including testis | |||
CO991452 PRDM7 susScr oviduct | |||
EW469934 PRDM7 susScr mucosal membrane | |||
The | The table below shows the number of zinc fingers in the second column, phylogenetic clade in the third, and adjacent gene (synteny) in the fifth. The number and character of zinc fingers is quite variable in human populations and likely so in all mammals; the table provides that of the individual selected for reference genome project which may not be representative of the species. | ||
These zinc finger arrays have been corrected in low coverage genomes for common sequencing errors -- frameshifts and premature stop codons arising from nucleotide run length mis-calls (eg, ggggg read as gggg) -- though they could actually represent valid mutant alleles in the heterozygous state (assuming the gene essential for meiosis). Indeed, these errors seem far more common than in what is seen in housekeeping genes for the same genome. | |||
The PRDM7 genes are all orthologous in the classical sense (as can be seen by adjacency to the unrelated gene GAS8) but various PRDM9 genes arose as different lineage-specific segmental duplications so are orthologous within a delimited phylogenetic clade. There is currently no suitable nomenclature for different gene duplications in different clades of the same parental gene so they are just called PRDM9 here, with PRDM7 reserved to genes adjacent to GAS8. In some species such as mouse, chromosomal rearrangements have scattered syntenic relations and orthology remains slightly uncertain but the single gene in the genome probably represents simple descent from the single euarchontoglire PRDM7 gene. | |||
* <font color=blue>PRDM7</font>: genes with ancestral location GAS8 synteny | |||
* <font color=green>PRDM9</font>: lineage-specific segmental duplications of PRDM7 | |||
* <font color=red>Pseudogenes</font>: multiple disabling frameshifts and stop codons in parental gene (not a retrogene) | |||
< | <font color=green> >PRDM9_homSap 13 prim gene CDH12 Homo sapiens (human) NM_020227</font> | ||
<font color=green> >PRDM9_panTro 19 prim gene CDH12 Pan troglodytes (chimp) GU166820</font> | |||
>PRDM9_homSap Homo sapiens (human) | <font color=green> >PRDM9_gorGor - prim gene cdh12 Gorilla gorilla (gorilla) CABD02290264</font> | ||
<font color=green> >PRDM9_ponAbe 10 prim gene CDH12 Pongo abelii (orangutan) XR_093432</font> | |||
<font color=green> >PRDM9_nomLeu 10 prim gene cdh12 Nomascus leucogenys (gibbon) ADFV01015315</font> | |||
<font color=green> >PRDM9_macMul 9 prim gene CDH12 Macaca mulatta (rhesus) XM_001083675</font> | |||
<font color=green> >PRDM9_papHam 11 prim gene cdh12 Papio hamadryas (baboon) genome</font> | |||
<font color=blue> >PRDM7_homSap 3 prim gene GAS8+ Homo sapiens (human) genome</font> | |||
<font color=blue> >PRDM7_panTro 2 prim <font color = red>pseu</font> GAS8+ Pan troglodytes (chimp) genome</font> | |||
<font color=blue> >PRDM7_gorGor 3 prim <font color = red>pseu</font> GAS8+ Gorilla gorilla (gorilla) genome</font> | |||
<font color=blue> >PRDM7_ponAbe 4 prim gene GAS8+ Pongo abelii (orangutan) genome</font> | |||
<font color=blue> >PRDM7_nomLeu 5 prim <font color = red>pseu</font> gas8+ Nomascus leucogenys (gibbon) ADFV01125891</font> | |||
2 | <font color=blue> >PRDM7_macMul 2 prim <font color = red>pseu</font> GAS8+ Macaca mulatta (rhesus) genome</font> | ||
<font color = | <font color=blue> >PRDM7_papHam 2 prim <font color = red>pseu</font> gas8+ Papio hamadryas (baboon) genome</font> | ||
<font color = | <font color=blue> >PRDM7_calJac 12 prim gene GAS8+ Callithrix jacchus (marmoset) XR_090591</font> | ||
<font color=blue> >PRDM7_tarSyr - prim <font color = red>pseu</font> gas8+ Tarsius syrichta (tarsier) ABRT011082008</font> | |||
<font color=blue> >PRDM7_micMur 8 prim gene gas8+ Microcebus murinus (lemur) ABDC01433247</font> | |||
<font color=blue> >PRDM7a_otoGar 10 prim gene GAS8+ Otolemur garnettii (galago) genome</font> | |||
<font color=blue> >PRDM7b_otoGar 8 prim gene GAS8+ Otolemur garnettii (galago) genome</font> | |||
<font color=blue> >PRDM7_tupBel 9 prim gene noDet Tupaia belangeri (tree_shrew) genome</font> | |||
<font color=blue> >PRDM7_oryCun 4 glir gene other Oryctolagus cuniculus (rabbit) genome</font> | |||
<font color=blue> >PRDM7_ochPri - glir gene noDet Ochotona princeps (pika) AAYZ01312269</font> | |||
<font color=blue> >PRDM7_ratNor 10 glir gene PDCD2 Rattus norvegicus (rat) NM_001108903</font> | |||
<font color=blue> >PRDM7_musMus 12 glir gene PDCD2 Mus musculus (mouse) NM_144809</font> | |||
<font color=blue> >PRDM7_musMol 11 glir gene noDet Mus molossinus (wild_mouse) GU216230</font> | |||
<font color=blue> >PRDM7_criGri 3 glir gene noDet Cricetulus griseus (hamster) AFTD01086355</font> | |||
<font color=blue> >PRDM7_dipOrd - glir gene noDet Dipodomys ordii (kangaroo_rat) genome</font> | |||
<font color=blue> >PRDM7_speTri - glir gene noDet Spermophil tridecemlin (squirrel) AAQQ01308561</font> | |||
<font color=green> >PRDM9a_bosTau 7 laur gene noDet Bos taurus (cattle) NW_003053109</font> | |||
<font color=green> >PRDM9b_bosTau 5 laur gene noDet Bos taurus (cattle) DAAA02065087</font> | |||
== | <font color=green> >PRDM9c_bosTau - laur gene noDet Bos taurus (cattle) XM_002699750</font> | ||
<font color=green> >PRDM9d_bosTau 9 laur gene noDet Bos taurus (cattle) genome</font> | |||
<font color=green> >PRDM9e_bosTau 9 laur gene noDet Bos taurus (cattle) genome</font> | |||
<font color=green> >PRDM9e_oviAri - laur <font color = red>pseu</font> noDet Ovis aries (sheep) genome</font> | |||
<font color=green> >PRDM9d_oviAri - laur gene noDet Ovis aries (sheep) genome</font> | |||
<font color=green> >PRDM9c_oviAri 4 laur <font color = red>pseu</font> noDet Ovis aries (sheep) genome</font> | |||
<font color=green> >PRDM9b_oviAri 2 laur <font color = red>pseu</font> noDet Ovis aries (sheep) genome</font> | |||
<font color=green> >PRDM9a_oviAri 9 laur gene noDet Ovis aries (sheep) genome</font> | |||
<font color=green> >PRDM9d_munMun 4 laur gene noDet Muntiacus muntjak (muntjac) AC216498</font> | |||
<font color=green> >PRDM9c_munMun 15 laur gene noDet Muntiacus muntjak (muntjac) AC154919</font> | |||
<font color=green> >PRDM9b_munMun 13 laur gene noDet Muntiacus muntjak (muntjac) AC218859</font> | |||
<font color=green> >PRDM9a_munMun 7 laur gene noDet Muntiacus muntjak (muntjac) AC225653</font> | |||
<font color=blue> >PRDM7_bosTau - laur <font color = red>pseu</font> GAS8+ Bos taurus (cattle) genome</font> | |||
<font color=blue> >PRDM7_turTru 9 laur gene gas8+ Tursiops truncatus (dolphin) ABRN01441536</font> | |||
<font color=blue> >PRDM7_lamPac 2 laur gene noDet Lama pacos (llama) scaffolds</font> | |||
<font color=blue> >PRDM7_susScr 9 laur gene GAS8+ Sus scrofa (pig) FP476134</font> | |||
<font color=blue> >PRDM7_canFam 5 laur <font color = red>pseu</font> GAS8+ Canis familiaris (dog) genome</font> | |||
<font color=blue> >PRDM7_felCat 11 laur gene GAS8+ Felis catus (cat) genome</font> | |||
<font color=blue> >PRDM7_ailMel 6 laur gene GAS8+ Ailuropoda melanoleuca (panda) GL193502</font> | |||
<font color=blue> >PRDM7_musPut 3 laur gene noDet Mustela putorius (ferret) AEYP01035077</font> | |||
<font color=blue> >PRDM7_neoVis 2 laur gene noDet Neovison vison (mink) JF288183</font> | |||
<font color=green> >PRDM9_pteVam 15 laur <font color = red>pseu</font> noDet Pteropus vampyrus (bat) ABRP01232219</font> | |||
<font color=blue> >PRDM7_pteVam 7 laur gene GAS8+ Pteropus vampyrus (bat) ABRP01250178</font> | |||
<font color=blue> >PRDM7_myoLuc 6 laur gene gas8+ Myotis lucifugus (bat) AAPE02062260</font> | |||
<font color=blue> >PRDM7_equCab 4 laur gene GAS8+ Equus caballus (horse) genome</font> | |||
<font color=blue> >PRDM7_sorAra 8 laur gene noDet Sorex araneus (shrew) AALT01000095</font> | |||
<font color=green> >PRDM9a_loxAfr 12 afro gene noDet Loxodonta africana (elephant) genome</font> | |||
<font color=green> >PRDM9b_loxAfr 3 afro <font color = red>pseu</font> noDet Loxodonta africana (elephant) genome</font> | |||
<font color=blue> >PRDM7_loxAfr 5 afro <font color = red>pseu</font> GAS8+ Loxodonta africana (elephant) genome</font> | |||
<font color=blue> >PRDM7_echTel 5 afro <font color = red>pseu</font> noDet Echinops telfairi (tenrec) genome</font> | |||
<font color=blue> >PRDM7a_proCap 17 afro <font color = red>pseu</font> noDet Procavia capensis (hyrax) ABRQ01392668</font> | |||
<font color=blue> >PRDM7b_proCap 13 afro <font color = red>pseu</font> noDet Procavia capensis (hyrax) ABRQ01227339</font> | |||
<font color=blue> >PRDM7_dasNov 9 xena <font color = red>pseu</font> noDet Dasypus novemcinctus (armadillo) AAGV020462211</font> | |||
<font color=blue> >PRDM7_choHof 2 xena <font color = red>pseu</font> noDet Choloepus hoffmanni (sloth) ABVD01893961</font> | |||
=== Gene trees based on domains === | |||
[[Image:PRDMcompBio.jpg|left]] | |||
PRDM9 is a chimeric protein consisting of 6 domains and linker regions. These domains occur in various combinations in many other human proteins without however known variability in domain order. The evolutionary relationships between all these proteins is necessarily complex, but taking the PR(SET) histone methylase as common denominator, the [[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2797608 gene tree] at left emerges after structural alignment considerations. | |||
While informative, this is really just a domain tree. A different tree would result based on the KRAB domain because it involves a different (though overlapping) set of proteins which had a partially independent history of duplication and shuffling from the PR(SET) domain. That precludes a meaningful joint tree based on KRAB + PR(SET) for those proteins that have both.The SSXRD domain has quite limited distribution but is considered further below. The knuckle and early zinc finger domains are rather short for domain tree inference, leaving presence/absence as the main consideration. | |||
A domain tree based on the terminal zinc finger array is problematic due to long independent histories of expansion and contraction. Here the main handle is the C2H2 classification (based not only on residues binding zinc but also their spacing). Main other types of zinc fingers occur in the human proteome. Some blur into C2H2 but others -- like the intertwined CCHC and HCCC in DRMT1 -- are structurally [http://www.ncbi.nlm.nih.gov/pubmed/10898790,8978051 quite distinct]. DRMT1 is the [http://www.nature.com/nature/journal/v461/n7261/full/nature08298.html sex-detemination gene in birds] and a [http://www.pnas.org/content/early/2010/07/02/1006243107.abstract major regulator of gene expression] in mammalian Sertoli and germ cells. It dramatically affects expression of mouse PRDM7 (called Prdm9) but apparently indirectly as the mouse gene lacks close-in upstream binding site according to genome browser [http://www.dmrt1.umn.edu/PNAS/index.php wig tracks]. | |||
The traditional PR(SET) domain seems too small for an enzyme with such distinctive substrates so [http://www.plosone.org/journals/journalNamePlaceholder/webapp/enhanced/pone.0008570/ flanking sequence] can be added consistent with observed amino acid conservation. Using S-adenosyl methionine as donor, PRDM9 places the third methyl group only on the fourth position lysine in mature histone H3 (which is actually position 5 prior to iMet removal: MART<font color =red>K</font>QTARK...), just one of this histone's [http://www.uniprot.org/uniprot/Q16695 27 modified residues]. There are many such epigenetic methylases in the human genome. PRDM9 has no applicable crystallographic structures, leaving undefined the residues involved in substrate binding and catalysis. | |||
The histone orthology class, methylation position and methylation extent of these methylases correlates poorly with evolutionary grouping by PR(SET) domain (figure), suggesting gene duplications can readily diverge in their properties. PR(SET) domains can even lose catalytic competence yet retain recognition capacity and recruitment of other proteins. However loss of constraints might lead to anomalously fast divergence and so to misplacement in the domain tree. | |||
PRDM7 | The upper left corner shows variability in domain structure. While PRDM9 and PRDM7 share the same domains (an upstream KRAB domain is not shown), of PR-class homologs, PRDM11 shares only the PR(SET) domain despite nesting deep within the PRDM9 subtree. PRDM4 has both the PR(SET) and C2H2 domains, possibly sharing the early zinc finger in an exon beginning with a phase 2 splice acceptor (marked up with color in reference sequence collection). Overall however, PRDM9 and PRDM7 have no full length homologs with matching exon structure. | ||
Note PR(SET) domain is even intronated differently within PR-class proteins, suggesting ancient divergence from a common pattern since intron gain/loss is exceedingly rare in vertebrates. These incongruities may have arisen from domain shuffling, gain and loss. Intron phasing provides a very important constraint on domain shuffling because the downstream reading frame must be preserved. | |||
Intronation patterns of PRDM9 domains fit the standard eukaryote pattern: domains evolved first, introns inserted later at random sites. Domain shuffling might be even more pervasive if domains corresponded cleanly with exon breaks and all introns were phase 0. However this is almost never the case, another instance of what is sometimes called 'unintelligent design'. | |||
The human PRDM9 sequence below is annotated in color for domains relative to exon breaks. The protein can be best understood in terms of concatenated domains, not all of which may be present in antecedent and descendant homologs. The first two domains KRAB and SSXRD likely interact with transcription factors, though these have not been specifically characterized in meiosis. | |||
Each terminal zinc finger type C2H2 array potentially recognizes a specific trinucleotide and so a large concatenated array quite specific binding sites in the genome, though tolerance of nucleotide variability and overlapping interactions between adjacent units make it difficult to read out these sites precisely, despite immense efforts. However aberrant individual zinc fingers are common in arrays; not all contribute directly to dna binding specificity. | |||
The concatenated C2H2 domains, conserved at the amino acid level so necessarily similar at the dna level, are apparently prone to replication slippage (or mis-registered gene conversion). Both processes can give rise to associated point mutations as well as leading to different repeat number distributions in human populations. | |||
Taking the extremes of variations, it is '''a wonder that humans can still interbreed''', yet [http://en.wikipedia.org/wiki/Haldane%27s_rule Haldane's Rule] so far has not been invoked as a cause of human infertility. PRDM9 allelic differences may provide an importent speciation barrier (ie, infertility of F1 males), yet introgression has been reported for denisovan, neanderthal and earlier African hominid dna into contemporary human genomes. By way of comparison, mice with 13 zinc finger repeats [http://www.sciencemag.org/lookup/resid/323/5912/373?view=full&uritype=cgi form sterile hybrids] when crossed with mice differing merely by an extra repeat. | |||
Many other unrelated proteins with internal repeats (such as the [[Coding_indels:_PRNP#The_peculiar_prion_repeat_expansion_in_Felids|octapeptide region]] of the prion gene PRNP) are also affected by replication slippage. These events, though rare, have been intensively studied because they cause toxic gain of function (Creutzfeldt-Jacob disease). Repeat expansions here too are accompanied by localized point mutation. The PRNP repeats have anomalously high GC content prone to self-similarity loop-outs unlike the C2H2 repeats of PRDM9. | |||
[ | Both PRDM9 and PRDM7 contain a seldom-mentioned zinc finger early in the final exon, as annotated by SwissProt and readily found by the online domain tools such as [http://smart.embl-heidelberg.de/ SMART] regardless of species. This domain conserves the four critical residues needed for zinc binding (and so the associated fold) but lacks the terminal cap TGEKP which otherwise serves to lock down a C2H2 zinc finger after it has scanned along genomic dna to an appropriate trinucleotide. The function of this early domain and the following 112 highly variable residues are unknown -- no demonstrably homologous sequence occurs in other human proteins with the possible exception of PRDM4 and PRDM15. | ||
The main zinc finger array also resides in this long distinctive terminal exon of splicing phase 12 that has been shuffled together into various contexts during mammalian evolutionary time. For once, intron phase is not so informative because the preceding PR(SET) domain with its codon overhang of 1 bp can accept any shuffled domain with overhang of 2 bp and still maintain reading frame. Since the KRAB domain also terminates in a phase 12 splice site, proteins can also skip the PR(SET) domain entirely, as in ZNF133 and many others. Concepts such as paralogy and orthology thus need piecewise definitions in these composite proteins. | |||
[ | The first C2H2 of the main repeat region is proximally degenerate, beginning in V<font color = blue>K</font>Y in all species (instead of Y<font color = blue>C</font>E). The lysine cannot plausibly replace the usual cysteine for zinc binding though the other three needed residues are present and may suffice. This domain ends in a typical cap region TGEKP. Humans are the exception here where the conserved helix-ending proline has been replaced with leucine in the reference human genome, with unknown functional consequences. This replacement is not recent since it is found in all human populations including the extinct [http://en.wikipedia.org/wiki/Denisova_hominin Denisovans] (41 kyr) and the basal (70 kyr) bushman lineage for which fragment V<font color = blue>K</font>YGECGQGFSVKSDVITHQRTHTGEK<font color = blue>L</font> YVCRECGRGFSWKSHLLIHQRIH is available from read 20_@FQ2QD2002IAZ67. | ||
< | |||
Another overlooked zinc finger domain occurs in the same exon as the PR(SET), preceding it. Being short, it is sometimes called a zinc knuckle rather than finger. There can be no doubt about its occurence because a crystallographic study has confirmed the [http://www.ncbi.nlm.nih.gov/pubmed/21604305 expected fold and zinc atom]. | |||
As noted, PRDM7 occurs immediately telomeric to the unrelated single-copy conserved gene GAS8 (with the two genes convergently transcribed). PRDM7 is otherwise the last gene on the q arm of its chromosome in many species which may predispose it to copy number dispersal events, which may in the past have resulted in juxtaposition and functional fusion to other genes. PRDM9 is not consistently present in placental mammals and each clade with it has a different syntentic location, suggesting numerous independent gene duplications (rather than many rearrangements and gene losses). | |||
[[Image:PRDM7dot.gif|left]] | |||
<br clear = all> | |||
>PRDM9_homSap Homo sapiens (human) Q9NQV7 10 exons chr5:23,509,579 span 18,301 bp <font color = green>KRAB</font> <font color = #00CC66>SSXRD</font> <span style="color: #990099;">zinc knuckle</span> <font color = #0066CC>PR(SET)</font> <font color = red>early ZNF</font> <font color = magenta>C2H2</font> <font color = blue>cap</font> | |||
0 MSPEKSQEESPEEDTERTERKPM 0 | |||
0 <font color = green>VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | |||
2 GLRATRPAFMCHRRQAIKLQVD</font>DTEDSDEEWTPRQQ 1 | |||
2 VKPPWMALRVEQRKHQK 0 | |||
0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 | |||
2 <font color = #00CC66>ELRKKETERKMYSLRERKGHAYKEVSEPQDDDY</font><span style="color: #990099;">L 1 | |||
2 YCEMCQNFFIDSCAAHGPPTFVKDSAV</span>DKGHPNRSALSLPPGL<font color = #0066CC>RIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 | |||
0 ITKGRNCYEYVDGKDKSWANWMR 2 | |||
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYG</font>QELGIKWGSKWKKELMAGR 1 | |||
2 EPKPEI<font color = red>HPCPSCCLAFSSQKFLSQHVERNH</font>SSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK | |||
<font color = magenta>VKYGECGQGFSVKSDVITHQRTH</font><font color = blue>TGEKL</font> | |||
<font color = magenta>YVCRECGRGFSWKSHLLIHQRIH<font color = blue>TGEKP</font> | |||
YVCRECGRGFSWQSVLLTHQRTH<font color = blue>TGEKP</font> | |||
YVCRECGRGFSRQSVLLTHQRRH<font color = blue>TGEKP</font> | |||
YVCRECGRGFSRQSVLLTHQRRH<font color = blue>TGEKP</font> | |||
YVCRECGRGFSWQSVLLTHQRTH<font color = blue>TGEKP</font> | |||
YVCRECGRGFSWQSVLLTHQRTH<font color = blue>TGEKP</font> | |||
YVCRECGRGFSNKSHLLRHQRTH<font color = blue>TGEKP</font> | |||
YVCRECGRGFRDKSHLLRHQRTH<font color = blue>TGEKP</font> | |||
YVCRECGRGFRDKSNLLSHQRTH<font color = blue>TGEKP</font> | |||
YVCRECGRGFSNKSHLLRHQRTH<font color = blue>TGEKP</font> | |||
YVCRECGRGFRNKSHLLRHQRTH<font color = blue>TGEKP</font> | |||
YVCRECGRGFSDRSSLCYHQRTH<font color = blue>TGEKP</font></font> YVCREDE* 0 | |||
-1 23 6 traditional numbering of dna recognizing amino acids | |||
LYCEMCQNFFIDSCAAHGPPTFVKDSAV alignment of zinc knuckle | |||
HPCPSCCLAFSSQKFLSQHVERNH alignment of pre-array zinc finger | |||
* * * * zinc liganding positions | |||
=== Segmental duplications creating PRDM9s from PRDM7 === | |||
[[Image:PRDM7segDup.gif|left]] | |||
In humans, PRDM9 and PRDM7 are related by a 26 kbp segmental duplication of chr16:90123419-90147718 that begins about 8 kbp upstream of the start codon and continues through most of the 3' UTR. Since the retroposon patterns are nearly identical, the duplication must be fairly recent. The overall percent identity of non-coding dna is about 93%, again inconsistent with either early (stem placental) or late divergence (great ape). The duplication contains a potentially diagnostic 1845 bp retroposon-free region upstream of the first coding exon. | |||
PRDM7 is situated at the extreme tip of chromosome 16q, perhaps predisposing it to chromosomal copy number variation (segmental duplications to other other chromosomes), ironically mediated by meiotic recombination. The syntenic context TUBB3+ DEFB+ AFG3L1+ DBNDD1- GAS8+ PRDM7- qTel means PRDM7 is transcribed convergently with GAS8, a non-homologous conserved single copy gene whose distal exons are often detectable even in low coverage genomes in the contig containing PRDM7. This association has been extremely stable over placental mammal evolutionary time and so serves to reliably distinguish PRDM7 orthologs from its spin-offs. | |||
The elephant genome has an old PRDM7 pseudogene adjacent to GAS8 in the expected opposite orientation. It has a second PRDM9-like copy in a novel syntenic location (ie unrelated to the CDH10-CDH12 location in primates) also seen in mammoth. Since afrotheres (plus xenarthrans) are the basal placental mammal, it follows that this locus too was spun off from PRDM7, establishing a 110 myr history of telomeric susceptibility of PRDM7 to repeated rearrangements. | |||
Gene copy variation may be common in individual inheritance but a paralogous copy seldom becomes established across a species and even more rarely displaces the parental gene completely. Yet this scenario is repeatedly observed in evolutionary history of PRDM7. Thus the telomeric location of PRDM7 may predispose it to these events, but their persistance is not accidental: the erasure of meiotic recombination iniation sites by biased gene conversion '''drives evolution of PRDM7/9 at three different scales''' -- point mutation at key amino acid positions, expansion/contraction of zinc finger tandem repeat number, and whole gene copy number. | |||
This history was not initially appreciated. Recall two genes in two species are orthologous only when they are vertically descended from the same gene in their last common ancestor which for human and elephant is post-marsupial mammal which had a single copy of PRDM7 adjacent to GAS8. The respective PRDM7 genes, still adjacent to GAS8 today are orthologous. The respective 'PRDM9' genes are however not descended from a common ancestral PRDM9 gene but from independent gene duplications of PRDM7 at different times during the course of afrothere and primate speciations. | |||
Human PRDM9 lies in a retroposon-rich gene desert, flanked by two pairs of cadherin genes at a larger scale of 7 mbp. In rhesus, these same genes are seen (with some minor rearrangements), by parsimony establishing this PRDM9 segmental duplication preceded the divergence of old world monkeys. | |||
Marmoset has a seemingly functional PRDM7 in the usual position facing GAS8, still at the extreme end of Callithrix chromosome 20. The cadherin cluster is intact on chr2:178,954,165-180,696,523. However Blastx of the intervening dna -- which is similar in size to rhesus and human so not suggestive of large deletions -- shows no suggestion of PRDM9. The assembly is gapless here. Blastx is sensitive enough to detect pseudogenes of this age provided they decayed only by small indels and nucleotide substitutions. | |||
Thus PRDM7 had not yet duplicated in the primate stem placing that event just prior to old world monkeys/great apes divergence. Note that the marmoset PRDM7 has a respectable terminal zinc finger array of twelve units, enough to specify an adequately specific dna recognition sequence. Tarsier assembly has poor coverage and only a fragmentary PRDM7 gene presumed adjacent to GAS8. | |||
Gene Strand Protein Start Species | |||
CDH18 - cadherin 18 19981287 homSap ponAbe macMul | |||
CDH12 - cadherin 12 22853731 homSap ponAbe macMul calJac | |||
PRDM9 + human PRDM9 23528704 homSap ponAbe macMul | |||
CDH10 - cadherin 10 24644911 homSap ponAbe macMul calJac | |||
CDH9 - cadherin 9 27038689 homSap ponAbe macMul | |||
Lemurs present a new complication. The Otolemur assembly has two distinct and possibly functional PRDM7 copies with 8-11 zinc fingers, according to how distal stop codons and frameshifts are interpreted in low coverage assemblies. One of these lies in a contig AAQR03144890 also containing GAS8 end-sequence in expected opposite orientation but this copy of GAS8 is a segmentally duplicated pseudogene, representing a new type of lineage-specific larger segmental duplication. Authentic GAS8 lies in a different Otolemur contig AAQR03166494 lacking any sign of a zinc finger protein. The second PRDM7 gene lies in a contig AAQR03189271 with novel synteny to the gene ARFGEF1. There is no sign of primate PRDM9 (a homolog intercalated between cadherins CDH10 and CDH12). | |||
The | The other lemur with an assembly, Microcebus murinus, has but a single presumptive PRDM7 with seven zinc fingers. The only relevant contigs (ABDC01433247 and ABDC01371462) contain no informative syntenic information so this gene cannot be associated with GAS8 with any confidence. However the contigs cannot be tiled and possibly belong to distinct genes. | ||
The basal euarchtonal species, tree shrew Tupaia belangeri, has an unsatisfactory assembly. A putative PRDM7/9 gene can be put together utilizing raw traces reads located with lemur blastn queries. These cannot be convincingly tiled and thus could originate from multiple genes including related chimeric domain proteins, even though best reciprocal blast of each exon calls up established PRDM7/9 matches. | |||
The | Moving on to laurasiatheres, Bos taurus presents a much more complicated situation. First, the GAS8 locus on chr18 contains the first two exons of a PRDM7 pseudogene in expected orientation but distal regions of the gene are completely deleted. The cadherin locus on chr20 is also intact but the 2.6 mbp region between CDH12 and CDH10 contains no indication of PRDM9, consistent with that segmental duplication being primate-specific and PRDM7 being the older parental location. This holds in the Baylor 4.0 assembly carried at UCSC, the Baylor 4.2 assembly, and the alternative assembly of the same data, UMD3.1. The latter two can be queried by the genomic [http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=9913 blast server] at NCBI. | ||
A third locus on chr 1 hosts an unreviewed GenBank pipeline entry called PRDM9, derived as NW_003053109 from the alternative bovine assembly UMD3.1. NCBI staff corrected an unspecified frameshift to fix the reading frame -- a dangerous practice in a gene family prone to pseudogenization. The gene, called PRDM9a here, resides on the extreme end of chromosome 1 and differs from the Baylor 4.0 assembly at two amino acids outside the zinc finger region. The syntenic context here is novel: EFHB- RAB5A+ PCAF+ ZNF596- PRDM9a- which corresponds overall to human chr 3. The juxtapositioning of two zinc finger proteins on the same strand causes PRDM9 alignments to extend spuriously into the 12 zinc fingers of ZNF596, jumping over its 5 earlier coding exons. | |||
ZNF596 contains a KRAB domain but no PR(SET) methylase. Humans encode a best-blast protein with the same assigned name on chr 8 (77% identity). Note the early exons of ZNF596 can be added to end of PRDM9a to form an artificial probe for this association in other species, though the two genes have a 43,400 bp spacer in cow, which is large relative to contig size in low coverage assemblies. The sole fragmentary transcript from yak testis (EF432551) is nearly identical to this PRDM9a, suggesting that the gene -- and perhaps its syntenic location -- became established prior to yak-cow divergence at 2.5 myr and is still functional. Its array of seven zinc fingers could recognize at most a region of 21 bp. | |||
ZNF596 did not arise from a PRDM9-like gene through loss of the SET domain, though it is one of the better matches within the large zinc finger family. Excluding the zinc finger domain, ZNF343, ZNF133 and ZNF169 provide much higher blastp scores, as they also do just comparing the zinc finger arrays. The juxtaposition of ZNF596 and PRDM9a is likely coincidental rather than a consequence of inhomogeneous recombination between zinc fingers bringing PRDM9 to this site. | |||
The fourth PRDM9 locus of interest, called here PRDM9b, is still not mapped to any bovine chromosome. It resides in contig DAAA02065087 in the UMD3.1 assembly and is temporarily assigned to chr Un.004.649 at Baylor assembly. Here the reading frame in exon two can be restored if a run of 5 A's is corrected to 6 A's, as homopolymer run length error is common in assemblies. The protein has a full set of domains KRAB SSXRD SET C2H2 with a moderate zinc finger array of five. Synteny cannot be determined in chr Un features which simple abuts unplaceable contigs into a manageable unit. Flanking dna in DAAA02065087 maps to several places in the cow genome, suggesting this feature has copy number attributes, perhaps of telomeric repeat type. PRDM9b is not a recent feature (or assembly stutte artifact) because it differs at toom any amino acids from other PRDM9 features in the cow genome. These substitutions avoid highly conserved residues, not consistent with cryptic pseudogenization. PRDM9b is capable of histone markup but it is not clear whether it does so. | |||
Yet another locus in the Baylor 4.0 assembly, called PRDM9c here, could not initially be placed on a cow chromosome. While such features are often assembly artifacts, this one is supported by a transcript from 4-cell embryos (GO353654) consistent with a role in or after meiosis. In UMD3.1, this gene has been placed on chr X. Despite a very large contig, no zinc fingers occur in any reading frame, suggesting that the gene was transferred here without the last exon or it subsequently got deleted. In any event, the penultimate exon does not have a phase 1 splice donor in expected position and so would terminate at the next stop codon downstream. The protein retains the KRAB, SSXRD and SET domains but does not possess the ability to scan or bind dna. It has accrued various amino acid substitutions relative to other bovine PRDM9 that rule out recent establishment. | |||
Finally, two additional genes, denoted PRDM9d and PRDM9e, are located as a parallel tandem pair in a higher quality region of bovine chr X. These are 96% identical as proteins, consistent with one being derived fairly recently from the other. Synteny here will not be informative until other ruminant genomes become available. | |||
Overall the situation in cow is very different from primates and rodents. Results there about the function of single-copy autosomal PRDM9 genes in meiosis markup can scarcely be carried over to a species with five seemingly intact genes, three of which are on chr X. Ruminants have a [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2593575/ well-characterized] small pseudoautosomal region on which crossovers with chrY can occur. | |||
The cow situation cannot be limited to the Hereford breed used for the genome project because the PRDM9 are too diverged from one another outside the zinc finger region. Indeed there is some suggestion from [http://www.livestockgenomics.csiro.au/blast/ non-NCBI sheep genome] that it too has many of these copies. Muntjak too seems similar to cow. However other cetartiodactyl genomes (dolphin, pig and alpaca) and other laurasiatheres (panda, dog, cat, shrew, bats) are not so expanded, suggesting that this unprecedented complexity could be limited to pecoran ruminants. The PRDM7 pseudogene is presumably parental to all these ruminant genes based on other laurasiatheres and placentals overall. | |||
All-vs-all blastp percent identities are consistent with this, though rates of evolution in this gene family hardly fit standard paradigms for quantification. Results for bovine are summarized in the table below: | |||
Gene #ZNF Status Chr Synteny cDNA Accession 9a_bosTau 9b_bosTau 9e_bosTau 9a_oviAri 9a_turTru 7_ailMel | |||
PRDM7 - pseudo 18 GAS8 no none -- -- -- -- -- -- | |||
PRDM9a 7 ok 1 ZNF596 yes NW_003053109 100% 85% 81% 82% 76% 72% | |||
PRDM9b 5 ok ? not det no DAAA02065087 81% 100% 78% 79% 72% 68% | |||
PRDM9c 0 ok X not det yes XM_002699750 80% 80% 82% 83% 74% 73% | |||
PRDM9d 9 ok X --- no none 80% 78% 96% 93% 73% 67% | |||
PRDM9e 9 ok X --- no none 81% 78% 100% 93% 73% 68% | |||
== Human PRDM9 variation == | |||
A great deal of attention -- and rightly so -- has been expended on cataloging variation in the zinc finger array at the level of both individuals and populations. While not the whole story of PRDM9 functionality by any means, this region is the primary determinant of recombination hotspot locations in meiotic dna. These sites greatly influence observed haplotypes and so the zinc finger array and its changing specificity over time must be understood to make reliable inferences about recent human evolutionary history and indeed speciation. | |||
The zinc finger array is roughly analogous to tRNA. Both bind trinucleotides, the former in double-stranded dna and the latter in single-stranded messenger rna. Both are somewhat fuzzy in binding specificity, the zinc fingers only partly specifying a sequence (eg CCNCCNTNNCCNC) and tRNA accepting wobble codons. Both require an array, these are covalently joined and consecutive in the zinc finger array but are discrete and sequentially acting in tRNAs. | |||
However this analogy only goes so far: the anticodons of tRNA have been fixed for billions of years whereas the four amino acid 'anticodons' of PRDM9 zinc fingers must undergo very rapid but highly restrictive mutation to keep up with an ever-changing recognition site (which obliterates itself with gene conversion, often the outcome of double-stranded break repair instead of recombination). Further, while all tRNAs recognize at least one codon, only a fraction of the zinc fingers in the human PRDM9 array can be utilized -- 13 fingers specify 39 nucleotides whereas observed sites are far shorter, some 13-17 base pairs. What selective pressure then maintains the unused fingers? | |||
That is but one of many remaining questions about PRDM9. Expression in some mammals is not restricted to germ line cells, suggesting other functionalities in the regulation of gene expression. The PRDM9 locus on chr5 itself does not contain a notable recombination hotspot (relative to its own zinc finger array) so gene conversion here cannot explain its mutational frequency, focus on the four determinative residues, and restricted compositional outcome (to nine of twenty amino acids). | |||
Selectional pressure on this gene is highly unusual in that an amino acid substitution in a germline cell yielding a zinc finger that cannot recognize a meiotic target is eliminated right away because recombination is essential to the meiotic process, meaning that no correctly divided haploid cell is available for fertilization. Other regions of the same protein evolve much more conventionally, with human PRDM9 diverging overall from other primates at unremarkable rates. | |||
The | The zinc finger array varies not only pointwise but also in number of repeats, from 13 or fewer to 20 or more, in contrast to many other stable 'polydactylic' zinc finger proteins. The mutational mechanism by which repeat numbers contract and expand has not been established but is presumably replication slippage, as in other unrelated proteins (such as the octapeptide repeat region in human PRNP). It is unclear what happens to individual zinc finger utilization after an expansion or contraction. | ||
Note in males, recombination must occur in the [http://en.wikipedia.org/wiki/Pseudoautosomal two short pseudoautosomal regions] of homology between chrX and chrY where few base pairs are available (relative to much longer autosomal chromosomes) for the recognition sequence to occur randomly with reasonable probability. Thus in humans PAR1 on the short-arm ends of chrX and chrY is 2.6 mbp whereas as PAR2 on the long arms ends only comprises 320 kbp. By comparison, the shortest human chromosome, chr22, has 50 million bases to host recombination recognition sites (16x as much). Thus the PARs may provide the do-or-die selectional bottleneck driving zinc finger array evolution. | |||
Given that small surveys in moderately inbred populations (such as Iceland) already find considerable variation in both number and sequence particulars of PRDM9 zinc finger arrays, it seems inevitable that many individuals must be heterozygous, sometimes radically so. However these would not necessarily be reported from sequencing projects where commonly only one allele is determined. It is not known whether both alleles in a heterozygous individual would be expressed and participate on an equal footing in meiosis in the same dividing cell. If so, the repertoire of recognizable sites would be expanded, with complications for understanding haplotype evolution if common. | |||
One last immense complication is that human and mouse do not speak for the rest of mammals. There, multiple copies are present in some major lineages, in some cases with zinc finger arrays too short to determine an adequately restrictive suite of recombination sites. Here the possibility must be considered that paralogous copies can act in tandem with short arrays acting in concert to define adequate length sites. The pseudoautosomal regions are by no means strictly conserved phylogenetically. Here adequate data may well be available from horse and cattle breeders but it has not surfaced to date. | |||
=== The role of CpG mutations === | |||
Human PRDM9 has 39 CpG sites in its coding exons, potentially methylated on the C, subject to spontaneous deamination to uracil and mis-repair, and so mutational hotspots. After attempted dna repair, the resulting change can be either CpA or TpG. These changes alter the encoded amino acid at non-synonymous sites. Some 28 of the CpG sites of PRDM9 are at arginine CGn codons (of which the protein has 90 overall). | |||
These always result in a substitution: G -> A mis-repair yields histidine for CGT and CGC and glutamine for CGG and CGA; C -> T mis-repair leads to cysteine for CGT and CGC and tryptophan and stop codon for CGG and CGA. These changes indeed occur in reported human and mammal sequences where they are perhaps best viewed as cSNPs in an individual rather than representing the species as a whole. The display below shows wildtype human PRDM9 in the top lines and the effects of G -> A and C -> T in the next. | |||
In terms of upstream CpG islands that would protect against methylation of CpG in coding regions, PRDM9 has none. While three occur somewhat near the start of PRDM7, these do not extend into coding exons and may not even be associated with this gene. The composite snapshot below from chr5 and chr16 of the UCSC human genome browser displays these CpG islands relative to the two genes. Thus CpG cytidines would be methylated in coding regions of both PRDM7 and PRDM9, rendering them susceptible to hotspot mutations. | |||
[[Image:CpGislandsPR.gif|left]] | |||
<br clear=all> | |||
In the terminal zinc finger array of the human PRDM9 reference sequence, position -1 is sensitive to the CpG hotspot effect. However rapid rapid evolution in the zinc finger array, which is overwhelmingly concentrated in the four dna-recognizing residues, cannot be explained by the CpG effect. On the other hand, the common alteration of the terminal partial finger YVCREDE* to Y*CREDE* in some species likely is a CpG effect but one that is insufficient for loss of function. | |||
PRDM9_homSapWT MSPEKSQEESPEEDTERTERKPM<font color=green>VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITIGLRATRPAFMCHRRQAIKLQV</font>DDTEDSDEEWTPRQQVKPPWMALRVEQRKHQKGMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL<font color=blue>ELRKKETERKM</font> | |||
PRDM9_homSapCA ...................Q.............................H...................Q......Q...................................H................................................................... | |||
PRDM9_homSapTG ...................W.............................C...................*......*...................................C........V.......................................................... | |||
PRDM9_homSapWT <font color=blue>YSLRERKGHAYKEVSEPQDDDYL</font>YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGL<font color=blue>RIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWLITKGRNCYEYVDGKDKSWANWMRYVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDE</font> | |||
PRDM9_homSapCA ...Q...........K........................................H.........................................Q....K......................................Q.....................Q............... | |||
PRDM9_homSapTG ...*............L.......................................C..............................L..........*...........................................W.....................*............... | |||
PRDM9_homSapWT <font color=blue>YG</font>QELGIKWGSKWKKELMAGREPKPEI<font color=brown>HPCPSCCLAFSSQKFLSQHVERNH</font>SSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK | |||
PRDM9_homSapCA .S..............................................H.....................................H............................................................................ | |||
PRDM9_homSapTG ................................................C.....................................C............................................................................ | |||
> | ........-1..23..6.......... ........-1..23..6.......... ........-1..23..6.......... ........-1..23..6.......... | ||
VKYGE<font color=blue>C</font>GQGSVKSDVIT<font color=blue>H</font>QRT<font color=blue>H</font>TGEKL YV<font color=blue>C</font>RE<font color=blue>C</font>GRGSRQSVLLT<font color=blue>H</font>QRR<font color=blue>H</font>TGEKP YV<font color=blue>C</font>RE<font color=blue>C</font>GRGRDKSHLLR<font color=blue>H</font>QRT<font color=blue>H</font>TGEKP | |||
.....<font color=blue>.</font>...........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.Q..Q......<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.Q.<font color=red>H</font>N......<font color=blue>.</font>...<font color=blue>.</font>..... | |||
.....<font color=blue>.</font>...........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.W..W......<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.W.<font color=red>C</font>.......<font color=blue>.</font>...<font color=blue>.</font>..... | |||
YV<font color=blue>C</font>RE<font color=blue>C</font>GRGSWKSHLLI<font color=blue>H</font>QRI<font color=blue>H</font>TGEKP YV<font color=blue>C</font>RE<font color=blue>C</font>GRGSWQSVLLT<font color=blue>H</font>QRT<font color=blue>H</font>TGEKP YV<font color=blue>C</font>RE<font color=blue>C</font>GRGRDKSNLLS<font color=blue>H</font>QRT<font color=blue>H</font>TGEKP | |||
.I<font color=blue>.</font>..<font color=blue>.</font>.Q.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.Q.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.Q.........<font color=blue>.</font>...<font color=blue>.</font>..... | |||
..<font color=blue>.</font>..<font color=blue>.</font>.W.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.W.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.W.........<font color=blue>.</font>...<font color=blue>.</font>..... | |||
YV<font color=blue>C</font>RE<font color=blue>C</font>GRGSWQSVLLT<font color=blue>H</font>QRT<font color=blue>H</font>TGEKP YV<font color=blue>C</font>RE<font color=blue>C</font>GRGSWQSVLLT<font color=blue>H</font>QRT<font color=blue>H</font>TGEKP YV<font color=blue>C</font>RE<font color=blue>C</font>GRGSNKSHLLR<font color=blue>H</font>QRT<font color=blue>H</font>TGEKP | |||
..<font color=blue>.</font>..<font color=blue>.</font>.Q.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.Q.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.Q.........<font color=blue>.</font>...<font color=blue>.</font>..... | |||
..<font color=blue>.</font>..<font color=blue>.</font>.W.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.W.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.W.........<font color=blue>.</font>...<font color=blue>.</font>..... | |||
YV<font color=blue>C</font>RE<font color=blue>C</font>GRGSRQSVLLT<font color=blue>H</font>QRR<font color=blue>H</font>TGEKP YV<font color=blue>C</font>RE<font color=blue>C</font>GRGSNKSHLLR<font color=blue>H</font>QRT<font color=blue>H</font>TGEKP YV<font color=blue>C</font>RE<font color=blue>C</font>GRGRNKSHLLR<font color=blue>H</font>QRT<font color=blue>H</font>TGEKP YV<font color=blue>C</font>RE<font color=blue>C</font>GRGSDRSSLCY<font color=blue>H</font>QRT<font color=blue>H</font>TGEKP YV<font color=blue>C</font>RE<font color=blue>D</font>E | |||
..<font color=blue>.</font>..<font color=blue>.</font>.Q..Q......<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.Q.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.Q.<font color=red>H</font>.......<font color=blue>.</font>...<font color=blue>.</font>..... .I<font color=blue>.</font>..<font color=blue>.</font>.Q..N......<font color=blue>.</font>...<font color=blue>.</font>..... .I<font color=blue>.</font>..<font color=blue>.</font>. | |||
..<font color=blue>.</font>..<font color=blue>.</font>.W..W......<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.W.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.W.<font color=red>C</font>.......<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>.W.........<font color=blue>.</font>...<font color=blue>.</font>..... ..<font color=blue>.</font>..<font color=blue>.</font>. | |||
A [http://weblogo.berkeley.edu/logo.cgi weblogo] based on alignment of placental mammal PRDM7 and PRDM9 genes (with pseudogenes excluded) illustrates the location of expected CpG mutations relative to conserved residues. These will be relatively high frequency loss-of-function alleles (not affecting health per se if only reproductive meiosis is affected). | |||
In the initial KRAB domain, the potentially affected arginines are not especially well-conserved. However, at the first site, neither histidine nor cysteine is part of the reduced alphabet so these changes are unlikely to be tolerated in meiotic functioning. At the second and third sites, glutamine does occur secondarily in some species (cow, sheep and muntjac) and murid rodents, respectively. These changes are thus borderline for adverse effects on functionality. | |||
[[Image:KRAB9logo.png|KRAB9logo.png]] | |||
<br clear=all> | |||
=== Sequence analysis of human variation === | |||
The PRDM9 terminal zinc finger array varies extensively in human, with significant consequences for hotspot recognition motif, distribution of recombination location options along the chromosomes, population history (linkage disequilibrium), and chromosomal rearrangement diseases. No other species -- notably other great apes -- has been surveyed to any extent for individual variation (with the exception of mouse PRDM7 where hybrid sterility was first mapped). | |||
For these species, we have only the sequence of the animal selected for genome sequencing and so have no idea whether human variation is unique or typical. With high priority chimp, Genbank contains only an uncurated erroneous gene prediction XM_517829 and an array fragment GU166820 with a disturbing number of differences to chimp reference genome. Gorilla is worse. Mouse has considerable variation in its zinc finger array but the strains involved are highly inbred and not necessarily representative of wild mouse diversity. | |||
Cheap short reads mapped to human reference as SNPs prove highly unsatisfactory for genes like PRDM9 where individuals differ not only at pointwise sites but also in wholesale repeat number. Several labs have reported novel repeat multiples but found an hour of re-sequencing too tedious; others assumed all possible arrays had already been reported and forced reads into one of these pre-existing classes; others left their discoveries as article graphics, behind firewall or in supplemental, not troubling themselves with GenBank entries, with [http://www.ncbi.nlm.nih.gov/nuccore/NM_020227,FJ899869,FJ899872,FJ899895,FJ899905,GU183914,GU183915,GU183916,GU183917,GU183918,GU183919,GU216222,GU216224,GU216225,GU216226,GU216227,GU216228,GU216229,HM210983,HM210984,HM210985,HM210986,HM210987,HM210988,HM210989,HM210990,HM210991,HM210992,HM210993,HM210994,HM210995,HM210996,HM210997,HM210998,HM210999,HM211000,HM211001,HM211002,HM211003,HM211004,HM211005,HM211006 laudable exceptions]. Even if certain arrays are rare, they provide invaluable information on the genetic mechanisms by which repeat number variation arises. | |||
[[Image:PRDM9gubi.gif|left]] | |||
It appears that few individual human genome or exome projects really gathered enough data to allow ab initio assembly of the zinc finger repeat array, or even when they did, walked away from that exercise, deposited a mess of indels and base miscalls at the Short Read Archive and then claimed SNPs relative to human reference, contaminating that resource with error. | |||
This is very unfortunate in the case of both basal and ancient human dna, which might record intermediate or population-specific stages in the evolution of human PRDM9. Extracting accurate bushman, paleo-eskimo, neanderthal, and denisova PRDM9 zinc finger arrays requires starting from scratch from raw read data. This may however be impossible due to inadequate coverage and confusion of short reads with PRDM7 and even within PRDM9, not to mention other closely related zinc finger proteins. | |||
Here PSU provides an [http://main.genome-browser.bx.psu.edu/cgi-bin/hgTracks excellent display] of reads (along with quality scores) reported by the various projects. The final exon of PRDM9 can be viewed (noting PSU uses hg18 coordinates) at chr5:23562098-23562523 for the early region and chr5:23562524-23563636 for the terminal zinc finger array. Viewing the display to dense mode shows the extent of tiling: it does not appear that adequate coverage was obtained in the critical cases. Here it cannot be assumed that the zinc finger array of bushman (who represent the earliest diverging living relative of Europeans) will closely resemble extensively sequenced West African variants (Yoriba). The best that can currently be done with bushman genome is VKYGECGQGFSVKSDVITHQRTHTGEKL YVCRECGRGFSWKSHLLIHQRIH ... YHQRTHTGEKP YVCREDE* which matches hg19 human reference sequence without shedding any light on internal repeat length or sequence variation. | |||
Although the zinc finger array conveniently resides in a single exon, that exon is almost never sequenced in its entirety. It has never been sequenced as a byproduct of an expression project. Consequently we have no idea its early zinc finger covaries with the terminal array nor any understanding of the constraints acting on the long bridging domain. | |||
10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 | |||
| | | | | | | | | | | | | | | | | | |||
PRDM9_homSap EPKPEI<span style="color: #0066CC;">HP<span style="color: #FF0000;">C</span>PSC<span style="color: #FF0000;">C</span>LAFSSQKFLSQ<span style="color: #FF0000;">H</span>VERN<span style="color: #FF0000;">H</span>SSQN</span>FPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK<span style="color: #0066CC;">VK<span style="color: #FF0000;">Y</span>GE<span style="color: #FF0000;">C</span>GQGFSVKSDVIT<span style="color: #FF0000;">H</span>QRT<span style="color: #FF0000;">H</span>TGEKL</span> | |||
PRDM9_panTro ...............................................................R................................................................A........................D.............G.P | |||
PRDM9_gorGor .........................................T.....................R.........................................................................................................P | |||
PRDM9_ponAbe .......................................................H....S..R.C.......................................................................................D.............GRS | |||
PRDM9_nomLeu ...A......................A.H.............F.................S..R.C...............................S..V...........I..-.............Q...........E............................ | |||
PRDM9_macMul ...............................T.........R.F....L.S.........S..R.C.....................PK................S...E.M....Y........E...........................D.....I.........P | |||
PRDM9_papHam ...............................T...R.....R......L.S.........S..R.C....................R.K................S...E.M...........S.E.I.........................D....VI.........P | |||
PRDM7_calJac .S.....................H.............T.T............K.KE....F..CNS........T........I......MA....N........S..EE.M.............VD...............A..........DM...TG.........P | |||
PRDM7_micMur ............S.............KHT....IS.RT.G..H................HS....C...A.D..V...P.PFH.K.Q..G...........K.....E........P......G..D.D..CAAG.....SR....DS..S..D..N..I.........P | |||
PRDM7_otoGar ............S....T..........T.P..ISQ.T.G..N.R.QT...R.E.....HS..N..........V..M..TSH.K.Q.SR...I..C........S.E.E.MI...P.PD...G..D.E.FC.AI...G.V...NR..V.S..N..N-LR.........P | |||
A '''terrible error''' was made early on in human PRDM9 variant nomenclature. It is completely unacceptable for PRDM9 to have a private system for naming zinc fingers that stands in conflict with dozens of previously established crystallographic structures, SwissProt, SCOP and PFAM practice, and nomenclature for the other 842 zinc fingers encoded in the human genome. | |||
Wrong as it is, the current nomenclature will not be easy to displace. However this site uses a nomenclature consistent with physical structure, comparative genomics and historical precedent throughout and only provides a partial translation table to the numerous misguided motif naming systems. | |||
The mistake arose because the first and last zinc fingers in primate PRDM9 are mildly anomalous. However it is exceedingly common even for internal zinc fingers to depart from canonical form, even to admit different spacings and substitutions in the C2H2 ligands, as well depart in length and cap domain. Zinc finger arrays commonly terminate in fragmentary motifs that often continue for a while in another reading frame (ie, represent non-3n indels with run-on to the next encountered stop codon). | |||
Even when each zinc finger is letter-perfect, only a small subset seem to function in dna recognition -- thus 15 zinc fingers in a PRDM9 variant could theoretically recognize a 45 bp dna sequence but a look at meiotic events show that 17 bp seems the upper limit for specificity, meaning no more than 6-7 motifs are utilized in vivo. Nomenclature must acknowledge all zinc fingers whether they are anomalous or not (or functional or not). | |||
Some 7,000 of the overall ten thousand zinc fingers end in a structurally distinct cap unit, typically TGEKP. This was shown long ago to lock the zinc finger down after scanning and has found the recognition sequence. Proteins with a single zinc finger still have this motif. It occurs at the end -- not the beginning -- of the main zinc binding region. Proline is no accident here: as a cyclic imino acid, it is structurally terminating for helix and sheet. | |||
In summary, zinc fingers begin 5 residues before the second zinc cysteine, not at the second cysteine as in the 'ABCD...' nomenclature. Human PRDM9 begins with a full length zinc finger, but with a lysine at position 2 replacing the usual branched aliphatic, a tyrosine at position 3 replacing the first cysteine and a leucine replacing the terminal proline: V<span style="color: #FF0000;">KY</span>GECGQGFSVKSDVITHQRTHTGEK<span style="color: #FF0000;">L</span>. These oddities became stably established in the theran ancestor 135 myr ago (though departures -- and even the expected residues -- are seen in some species). Otherwise, the first zinc finger is quite conventional. The terminal leucine surprisingly is seen in all reported human variants. While the first zinc finger assuredly has the zinc finger fold and likely binds zinc to some extent, it likely does not function directly in specific dna motif recognition. Its role may more that of a macro cap, facilitating the lineup of downstream zinc fingers. | |||
Similarly, the terminal YVCRE<span style="color: #FF0000;">DE</span>* fragment, with its anomalous charged aspartate and glutamate in place of cysteine and glycine, is not zinc binding or part of a dna recognizing motif but simply a partial end cap. It has persisted (imperfectly) since the boreoeutheran ancestor so evidently provides significant value. In many laurasiatheres it is YCRECE or even YRCREG. Even a canonical hexapeptide cannot reach across the preceding TGEKP cap to displace zinc binding residues of the last full repeat, nor can the six residues circle around to displace the first five residues of first repeat. Instead, these residues form the start of an additional fold, enough to that keep the repeat array from unraveling. | |||
Below the available diversity at GenBank is shown, with the 0th GEKL repeat removed because it has no variation. Also the longest allele has its last two repeats removed to shorten the display width. The terminal fragment is also not shown. Redundant sequences have been largely removed from the set of 42. The alignment is shown at the protein level because synonymous dna variation is largely irrelevant to function. | |||
[[Image:Prdm9HumVarAlign.gif|left]] | |||
<br clear=all> | |||
If all [http://www.ncbi.nlm.nih.gov/nuccore/NM_020227,FJ899869,FJ899872,FJ899895,FJ899905,GU183914,GU183915,GU183916,GU183917,GU183918,GU183919,GU216222,GU216224,GU216225,GU216226,GU216227,GU216228,GU216229,HM210983,HM210984,HM210985,HM210986,HM210987,HM210988,HM210989,HM210990,HM210991,HM210992,HM210993,HM210994,HM210995,HM210996,HM210997,HM210998,HM210999,HM211000,HM211001,HM211002,HM211003,HM211004,HM211005,HM211006 42] human variant zinc finger array alleles at GenBank are collected, parsed into their zinc fingers and aligned for their differences relative to the genomic reference sequence, 25 variant fingers emerge at varying frequencies of occurrences. These are provided below, ordered by subgroup. The full sequence, allele name of the original investigators and representative accession are also given. | |||
As previously observed, variation at the amino acid level is overwhelmingly concentrated at that handful of internal positions recognizing dna bases in the major groove. Furthermore, the amino acid substitutions are strongly concentrated within only 9 of the 20 available amino acids. These observations raise the question of how ordinary random mutational processes could possibly have produced these results. Perhaps variation elsewhere results in failed meiosis, causing these to disappear immediately, leaving only the observed variation. | |||
Note in the table below (whose underlying data is [http://genomewiki.ucsc.edu/index.php/File:ZNFvariation.pdf here]) that threonine appears as a very common alternative to isoleucine outside the critical region (between the two zinc-binding histidines): YVCRECGRGFSWKSHLLIHQR<span style="color: #FF0000;">I</span>HTGEKP. This is actually an oddity of the first GEKP repeat: T is found here in all other primates (ie ancestrally), not I. As bushmen also have I here, this allele was fixed prior to their divergence at 70,000 years. | |||
Human Variation in 507 Zinc Fingers in 42 PRDM9 Variants | |||
Difference to NM refSeq Freq Full length zinc finger Accession Allele Great Ape Zinc Finger Variation | |||
> | YVCRECGRGFSWKSHLLIHQRIHTGEKP 39 YVCRECGRGFSWKSHLLIHQRIHTGEKP NM_020227 ref YVCRECGRGFSWKSHLLIHQRIHTGEKP homSap | ||
......................R..... 1 YVCRECGRGFSWKSHLLIHQRIRTGEKP FJ899869 7 .................S...T...... 1 panTro | |||
............Q.V..T...T...... 100 YVCRECGRGFSWQSVLLTHQRTHTGEKP NM_020227 ref ...........V..S..S...T...... 6 panTro | |||
............Q.V..S...T...... 22 YVCRECGRGFSWQSVLLSHQRTHTGEKP GU216222 A ...........V..S..S.RTT...... 1 panTro | |||
............Q.V..R...T...... 13 YVCRECGRGFSWQSVLLRHQRTHTGEKP GU183919 CH3 ...........VQ.N..S...T.....L 1 panTro | |||
......R.....Q.V..T...T...... 1 YVCRECRRGFSWQSVLLTHQRTHTGEKP HM211000 L18 ...........QQ.N..S...T...... 1 panTro | |||
............Q.VP.T...T...... 1 YVCRECGRGFSWQSVPLTHQRTHTGEKP FJ899895 18a ...........RQ.A......T...... 1 panTro | |||
...........N.....R...T...... 65 YVCRECGRGFSNKSHLLRHQRTHTGEKP NM_020227 ref ......E....QQ....R...T...... 1 panTro | |||
..........RN.....R...T...... 39 YVCRECGRGFRNKSHLLRHQRTHTGEKP NM_020227 ref ...........QQ....R...T...... 2 panTro | |||
..........RK.....R...T...... 1 YVCRECGRGFRKKSHLLRHQRTHTGEKP GU183915 AA2 ...........QQ....S...T...... 2 panTro | |||
..........RD.....S...T...... 14 YVCRECGRGFRDKSHLLSHQRTHTGEKP GU216229 I ...........KQ....S...T...... 2 panTro | |||
..........RD.....R...T...... 27 YVCRECGRGFRDKSHLLRHQRTHTGEKP NM_020227 ref ...........RQ.V......T...... 1 ponAbe | |||
..........RD..N..S...T...... 48 YVCRECGRGFRDKSNLLSHQRTHTGEKP NM_020227 ref ...........RR.V......T...... 1 ponAbe | |||
..........RD..P..S...T...... 1 YVCRECGRGFRDKSPLLSHQRTHTGEKP GU183915 AA2 ...........QQ.V......T...... 1 ponAbe | |||
..........RD..N..S...T...D.. 4 YVCRECGRGFRDKSNLLSHQRTHTGDKP GU183915 AA2 ...........RR.V......T...... 1 ponAbe | |||
..........RDE.N..S...T...... 2 YVCRECGRGFRDESNLLSHQRTHTGEKP HM211006 24L ..............V..R...T...... 1 ponAbe | |||
..........RDQ....S...T...... 1 YVCRECGRGFRDQSHLLSHQRTHTGEKP GU183919 CH3 ...........QQ.VVF....T...... 1 ponAbe | |||
...........RQ.V..T...T...... 2 YVCRECGRGFSRQSVLLTHQRTHTGEKP FJ899905 10b ...........G..V.FR...T...... 1 ponAbe | |||
...........RQ.V..T...R...... 79 YVCRECGRGFSRQSVLLTHQRRHTGEKP NM_020227 ref ...........D..GVCY...T...... 1 ponAbe | |||
...........RQ.V..T...G...... 2 YVCRECGRGFSRQSVLLTHQRGHTGEKP FJ899872 10 ...........V..N..S...T..E..L 1 ponAbe | |||
...........RQ.V..S...T...... 1 YVCRECGRGFSRQSVLLSHQRTHTGEKP GU216228 H ...........D..S..R...T...... 3 nomLeu | |||
...........NQ.V..T...T...... 1 YVCRECGRGFSNQSVLLTHQRTHTGEKP GU183916 AA11 ...........K..N..S...T...... 1 nomLeu | |||
...........DQ.V..T...T...... 1 YVCRECGRGFSDQSVLLTHQRTHTGEKP GU183916 AA11 ...........V..N..S...T...... 1 nomLeu | |||
...........DR.S.CY...T...... 37 YVCRECGRGFSDRSSLCYHQRTHTGEKP HM210983 L1 ...........Q..S..S...T...... 3 nomLeu | |||
...........DR.S.CY...T..MSKS 5 YVCRECGRGFSDRSSLCYHQRTHTMSKS GU183916 AA11 .L.........V..S..S...T...... 1 nomLeu | |||
507 | |||
When the 42 variants are aligned at the dna level, synonymous variation might be anticipated more or less evenly across the repeat array under the assumption that natural selection acts here only at amino acid level. However this is not the case as shown in the graphic below. For example, the GEKL repeat has no variation whatsoever despite numerous 4N codons. Elsewhere, synonymous variation is again highly concentrated at residues important to meiotic repeat recognition. This suggests a novel mutational mechanism exists that focuses change at the key regions, at a rate far above the genomic average. Conceivably the dna itself might have additional hairpin structure that exposes the critical regions to enhanced mutation. Such a speculative structure would fit with replication slippage varying the number of array repeats. This mechanism can also sweep out variation. Alternatively, the observed distribution of synonymous variation could arise via hypothetical mRNA editing in conjunction with a retroposon-like or copy-editing mechanism. A third option envisions another protein recognizing the dna encoding the repeats and acting upon them to provide variation. | |||
[[Image:PRDM9syn.gif]] | |||
The terminal zinc finger array in the human reference sequence (but not chimp or orangutan) has a two-block structure. That is, the repeats 3-7.5 have a high degree of internal self-similarity as do repeats 7.6-13.4. However these two blocks are markedly dissimilar to each other, primarily due to transversions (rather than transitions C<->T or A<->G). The genetic code is such that transversions give rise to markedly less conservative amino acid substitutions in both physical and dna binding properties, as can be readily seen at the protein level for human PRDM9. | |||
The origin of the two-block feature cannot be dated accurately because of limited sampling of individuals in great apes but is likely specific to human (or even to some human populations). It is closely correlated with the history of repeat contraction and expansion in this rapidly changing region of the gene. Note the zinc finger array is neither a [http://en.wikipedia.org/wiki/Microsatellite_%28genetics%29 microsatellite] being 84 bp long vs the requirement of 1-6 bp nor a [http://en.wikipedia.org/wiki/Minisatellite minisatellite]. | |||
[http://www.vivo.colostate.edu/molkit/dnadot/ Dotplots] can create visual artifacts, depending on scanning window and mismatch settings. Here the two-block structure is highly robust to exploration of parameter space and its basis is readily apparent at both the dna and protein sequence levels. | |||
[[Image:Prdm9Blocks.gif]] | |||
<br clear = all> | |||
A very recent [http://www.pnas.org/content/108/30/12378.full.pdf+html?with-ds=yes PNAS article] looks at many human meioses and assigns the recognition sequence to the distal region of the zinc finger array (in Fig 1D and supplemental S2), which corresponds to the second block identified above. That raises the question whether the two blocks evolve by different non-mixing mutational mechanisms and leaves unexplained the functional tasks implied by observed conservation of the first block. The lack of block structure in chimpanzee PRDM9 illustrates once again that meiotic initiation is evolving in many different directions. | |||
[[Image:BergRecogn.gif|left]] | |||
<br clear = all> | |||
=== Rate of proximal PRDM9 evolution in primates === | |||
The rate of evolution of human PRDM9 at the protein level -- excluding special evolution in the terminal zinc array -- is rapid but perhaps not unusually so. A rate anomaly can only be defined relative to the rather skewed rate distribution of the human proteome (20,000 loci). However a better comparison might be just to rates of the many other KRAB, SSXRD and PR(SET) domains in the proteome and to linker regions which are often under little selection. These regions have mediocre conservation in general and so a rapid rate there for PRDM9 has no immediate implications for its association with meiosis or protein binding partners. | |||
The fact that PRDM9 is a recent gene duplicate of PRDM7 adds another rate complication, as gene duplicates often exhibit rapid initial evolution as the copies subfunctionalize, a problem exacerbated here by functional persistence of a variously truncated PRDM7 in some primate lineages. Independent duplications of PRDM7 (eg afrotheres and pecorans) further complicate rate considerations outside of primates. | |||
Together, these considerations make it difficult to define a meaningful 'peer group' in any major clade by which to benchmark the rate of PRDM7/9 evolution. However by any measure this gene family is not evolving slowly in placental mammals, perhaps surprising because excluding the zinc finger array leaves domains with seemingly fixed and demanding functionality. That is, the KRAB domain is co-evolving with its protein binding partners which are under many other constraints. The histone substrate for methylation is exceedingly conserved but the PR(SET) catalytic domain of PRDM9 is not, despite its narrow specificity. | |||
The difference alignment below shows change localization relative to human functional domains within other primate PRDM9. The comparison includes catarrhine PRDM7 corrected where needed for frameshifts and stop codons as well as PRDM7 from euarchontal species that diverged before the gene duplication. The 532 residues are relatively free of deletions or insertions. | |||
None of the four sites where human diverges from long-established consensus (R5K, P155S, G178R, R445H) are CpG hotspots. With the exception of G178R, these are conservative substitutions in linker regions and likely near-neutral. G178R represents a radical change in amino acid properties within the SSXRD domain. However this is [[#Reciprocal_translocation:_origin_of_the_SSX1-PRDM_chimera|not a conserved residue]] in the parental SSX1 domain. | |||
Sequencing accuracy is an issue here because some genes are missing exons altogether and other exons have only single trace coverage. Outside of human and mouse, only a single individual has been sequenced. However humans are not especially variable in the proximal region of the gene, only a single coding SNP is known to date (R113C), in marked contrast to the zinc finger array. Much more intensive sequencing of primates is essential to quantitative understanding recent evolution or PRMD9 -- the 16 species sampled so far represent [http://en.wikipedia.org/wiki/Primate only 5%] of living primate diversity. For example, the flying lemur divergence node is not represented at all. | |||
<------------------------ KRAB domain ------------------------> cSNP:R113<font color=blue>C</font> <-------- SSXRD domain ---------><-------- zinc knuckle ----> <--------------- | |||
<font color=blue>PRDM9_homSap MSPEKSQEESPEEDTERTERKPMVKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITIGLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQVKPPWMALRVEQRKHQKGMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKLELRKKETERKMYSLRERKGHAYKEVSEPQDDDYLYCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWN | |||
PRDM9_panTro ....R...................................................................................................L.................................................P................K.....G.................................................................................... | |||
PRDM9_gorGor ....R...................................................................................................C.................................................P......................G............................................I....................................... | |||
PRDM9_ponAbe ....R.......D.........T.....................................................................................................N.........E...................P.................S....GNT.............I.........-....................................T..................... | |||
PRDM9_nomLeu ....R..............Q..T......................................................................................M........................GA..................P.................R....G.....<font color=black>*</font>............................T...........I...T.G............................... | |||
PRDM9_macMul ....R.................T....................................................................................V....S...........N.......V.GM.....T............P...R.............R....G..............................................I.....E............................... | |||
PRDM9_papHam ----------------------------------------------------------------------------------------------------.......F....S....E...T............G.P...ST.........A..P.................R..A.G..................L.................................N............................... | |||
</font><font color=brown>PRDM7_homSap ....R.......G..........................................M.......V...........................................F.G..S...........N.....R...G.P....T.D..........P.................R....G...............I.................................................................... | |||
PRDM7_panTro ....R...................................................................................................L.................................................P................K.....G.................................................................................... | |||
PRDM7_gorGor ....R.......G........................................................Q.V............................-----------------.......N.........G.P....T............P...........R.....R....G...............I.K....................................R............................. | |||
PRDM7_ponAbe ....R......KG...........................T.................KT...............................................F.G..S...........N.........G.Q....T............P..........T..I...R....G.T.................................................................................. | |||
PRDM7_nomLeu ----------------------------------------------------------------------------------------------------I.S....V....S...........N...G.....GSQ....T..<font color=black>*</font>...R.....P...........Q.....R....G.....<font color=black>*</font>............................T.......................H......................... | |||
PRDM7_macMul ----------------------------------------------------------------------------------------------------.......F....S....E...T..N.........G.P...ST.D..<font color=black>*</font>....A..P.................R....G.........R.....A..L.H............................N..N............................... | |||
PRDM7_papHam ....R..............W.......................................................................................V....S...........N.......V.GM.....T............P...R.............R....G..............................................I.....E...............................</font> | |||
<font color=green>PRDM7_calJac ....R.......G..G...Q............M..S.................M..................................................G..F..G.S...........G......K..G...V..T..P.........P.................R.D..E..........L.................I.............................HA........................ | |||
PRDM7_tarSyr ...DR.P.D...G..G...C.SA.........................I..........T..A.....P........KR...PL.......................F....N.......R.PL.IV.......EM....<font color=magenta>^</font>T.D....W......<font color=magenta>^</font>.....E....K.I.F....I.VN........DC.....N...........Q..........T..I...IN.................................... | |||
PRDM7_micMur ...N........V.AG..GW..TD...........S.....Q......I..........V........P.............H.................----------------------------------------------------------------------------------------------............K.......................................K.R............. | |||
PRDM7_otoGar ----------------------------------------------------------------....P...........T.YK..................H....F.M..S.R..ILK.CML.FNMH.....GP.S.P.I.....H..HM.SPR.........GR.SD..I..I.VR...........................K.......N..V.........T..E.......V....S..G.RT.......F....</font> | |||
>PRDM9_ponAbe Pongo abelii (orangutan) | ---------------------------- PR(SET) domain -------------------------------------------------------> <- early zinc finger --> | ||
<font color=blue>PRDM9_homSap EASDLPLGLHFGPYEGRITEDEEAANNGYSWLITKGRNCYEYVDGKDKSWANWMRYVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGI<font color=magenta>k</font>WGSKWKKELMAGREPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK | |||
PRDM9_panTro ..............K.......................................................................................................................................................................R................................................................A............. | |||
PRDM9_gorGor ..............K.................................................................................................................................................T.....................R.............................................................................. | |||
PRDM9_ponAbe ...................K..........................................................................................................................................................H....S..R.C............................................................................ | |||
PRDM9_nomLeu ..................................................................................Q.......P........................T.E....A......................A.H.............F.................S..R.C...............................S..V...........I................Q...........E | |||
PRDM9_macMul ...................Q..................................................................................................................................T.........R.F....L.S.........S..R.C.....................PK................S...E.M....Y........E................ | |||
PRDM9_papHam .....................K.................................----------------------------------------------------------------...............................T.........R......L.S.........S..C.C......................K................S...E.M.............E.I.....E........ | |||
PRDM7_homSap ..........................S......................S............................................S.................................................................................R..S..RCC.......................................S...E.M.............................. | |||
</font><font color=brown>PRDM7_panTro ..............K...................................................................................................................................................-.....<font color=black>*</font>.......R..S..RCC........V.........W............L.......S...E.M.............................. | |||
PRDM7_gorGor .....................K....S...........................................L............................................T............................................................R..S..RCC..................<font color=black>^</font>....<font color=black>^</font>...............S...E.M.............................. | |||
PRDM7_ponAbe .....................K......................................W.......................................................P...........................................H...A..............S..DCC...............................SA......S...E.M.....G........................ | |||
PRDM7_nomLeu .................................................<font color=black>*</font>....K.......H...................Q.......P........................T.E...............V.T..........C.................R..............S.SR.C...............-...I...K.......L.......S...E.M.............................. | |||
PRDM7_macMul .............C.......K....S........................K...----------------------------------------------------------------......Y................<font color=black>^</font>....................S..............S..S.C.........................L...T.........S...E.M....F.....A............E...... | |||
PRDM7_papHam ...................Q.........................................................................................................Y......................................S..............S..S.C....................R..Q.L...T.........S...E.M.K..F.....A............E......</font> | |||
<font color=green>PRDM7_calJac .................V.......SS.............................................................................................S.....................H.............T.T............K.KE....F..CNS........T........I......MA....N........S..EE.M.............VD............... | |||
PRDM7_tarSyr ...E............Q..D......S........................................................I....................................T..K.L..S..S..L.....F....KC..PP.I...T....YV.......E.L.....QS....W...S.C..A.....PMH...Q...S..SL.N..TE.TE.S.EKE.M.K..PS.S...HL.D..YE.HHI.A.AAR- | |||
PRDM7_micMur ...E............QV........S....................D..............E...................Q.............................E..TIRQ............S.............KHT....IS.RT.G..H................HS....C...A.D..V...P.PFH.K.Q..G...........K.....E........P......G..D.D..CAAG.....SR | |||
PRDM7_otoGar .....Q..........QV........S....................E.QG...........E....................................................T..Q............S....T..........T.P..ISQ.T.G..N.R.QT...R.E.....HS..N..........V..M..TSH.K.Q.SR...I..C........S.E.E.MI...P.PD...G..D.E.FC.AI...G.V- | |||
</font> | |||
As expected, percent identity declines monotonically with increasing time of divergence. The rate of amino acid substitution, subject to the caveats above, places PRDM9 in the lowest quartile of human protein conservation, but with thousands of proteins evolving still faster. Variability is noticeably but not exclusively concentrated in the linker between the KRAB and SSXRD domains and in the long terminal linker. Variation within the PR(SET) domain largely avoids deeply conserved residues defined from [[#Structural_alignment_of_all_PRDM_proteins|comparison]] of the 16 distinct PRDM genes and 35 additional SET domains in human. | |||
The table below shows percent amino acid difference relative to human, first for the entire 523 residues preceding the array and then separately for the three domains. Note the KRAB and PR(SET) are evolving slower than the overall proximal region while the SSXRD domain is changing more rapidly (however it is short so subject to wide rate swings). | |||
Missing exons were supplied here by merging incomplete sister taxa (lemurs) or taking them from the closest source (gibbon, gorilla, tree shrew PRDM7). Taken human/tree shrew divergence at [http://genomewiki.ucsc.edu/index.php/Phylogenetic_Tree 90 million years], roughly one substitution per million years has been occurring, much of it in the last exon between the early zinc finger and terminal array. | |||
In summary, the proximal region of PRDM9 is evolving quite differently than the zinc finger array. It is difficult to distinguish here between adaptive change, neutral drift, and substitutions driven by the role of PRDM9 in meiosis and recombination. | |||
KRAB SSXRD PR(SET) | |||
100% 100% 100% 100% PRDM9_homSap Homo sapiens (human) | |||
98% 100% 93% 99% PRDM9_panTro Pan troglodytes (chimp) | |||
98% 100% 96% 99% PRDM9_gorGor Gorilla gorilla (gorilla) | |||
96% 100% 84% 99% PRDM9_ponAbe Pongo abelii (orangutan) | |||
94% 100% 90% 98% PRDM9_nomLeu Nomascus leucogenys (gibbon) | |||
94% 100% 93% 99% PRDM9_macMul Macaca mulatta (rhesus) | |||
94% 96% 90% 97% PRDM7_homSap Homo sapiens (human) | |||
96% 100% 93% 99% PRDM7_panTro Pan troglodytes (chimp) | |||
94% 96% 87% 97% PRDM7_gorGor Gorilla gorilla (gorilla fusion) | |||
93% 95% 90% 98% PRDM7_ponAbe Pongo abelii (orangutan) | |||
91% n/a 90% 95% PRDM7_nomLeu Nomascus leucogenys (gibbon fusion) | |||
93% 100% 93% 99% PRDM7_papHam Papio hamadryas (baboon) | |||
90% 95% 87% 97% PRDM7_calJac Callithrix jacchus (marmoset) | |||
80% 87% 78% 95% PRDM7_tarSyr Tarsius syrichta (tarsier) | |||
81% 90% 84% 92% PRDM7_micMur Microcebus murinus (lemur fusion) | |||
73% 76% 75% 92% PRDM7_tupBel Tupaia belangeri (tree shrew) | |||
=== Variation in closely related ZNF proteins === | |||
Among the 843 human genetic loci encoding zinc fingers proteins, the arrays most closely resembling PRDM9 in length, structure and amino acid composition are ZNF133, HKR1, ZNF343, ZNF589, ZNF169, ZNF596. While the functions of these proteins are largely unknown, the first two have a KRAB domain, a spacer, early zinc finger in the terminal phase 2 exon, and a zinc finger array similar in size to human. The next two are similar but lack the spacer, with the KRAB domain encroaching into the final exon. The final two have only the KRAB domain and terminal array. Some 290 human gene products encode a KRAB domain. | |||
Here ZNF133 and the misnamed HKR1 are the best candidates for donating (via inhomogeneous recombination) the zinc finger array to the nascent PRDM7 which was already a chimer of KRAB, SSXRD and PR(SET) domains. The relationships here might instead go the other way (domain loss in PRDM) but different intronation of the KRAB domain is incompatible with that scenario. While none of the six ZNF is capable of histone methylation, KRAB domains are [http://www.ncbi.nlm.nih.gov/pubmed/11959841 capable of recruiting] SETDB1, a H3K9 methylase, partnering with the TIF1ß co-repressor protein (encoded by TRIM28), which interacts with many KRAB domains). | |||
Phylogenetic variation in the zinc finger arrays of these proteins is potentially quite informative, the question being whether their variation too is focused on the four amino acid positions providing dna binding specificity in PRDM7/9. This next sections examine each protein separately for mutational variation in the zinc fingers over placental mammal evolutionary time. | |||
Here the [http://genome-test.cse.ucsc.edu/cgi-bin/hgPal?g=knownGene&c=chr20&l=18269121&r=18297638&i=uc010gcs.2&hgsid=2970269&db=hg19 46-species genomic alignment] at UCSC serves as initial source of zinc finger arrays, which are then tested by blat back into individual species and then parsed into separate fasta files for each protein finger (the formats needed by the [http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_multalin.html Multalin2] variable width differential aligner, [http://weblogo.berkeley.edu/logo.cgi weblogo] and [http://www.vivo.colostate.edu/molkit/dnadot/ DotPlot tool]). | |||
==== ZNF133 and HKR1 ==== | |||
Human ZNF133 is a conventional KRAB-zinc finger array (that lacks however the PR(SET) domain). Although the KRAB domains are only 31% identical, the array provides a better model for PRDM9 than the other 14 PRDM* loci in terms of zinc repeat character and length. However rodents cannot be used here as a model system for ZNF133 as the mouse syntenic counterpart is a [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2922377/ known pseudogene] -- as is rat but not guinea pig or rabbit. ZNF133 is yet another protein in this class that does not readily track back into marsupials or earlier vertebrates. | |||
As with PRDM7/9, the C-terminal run-off of ZNF133 is subject to frameshifts. However elephant and human are still 86% identical in their last exon, with zinc finger arrays even higher. Armadillo, another mammal diverging from human at 101 myr, is 91% identical in this region and has exactly the same number of zinc fingers (14.7). This suggests that the dna binding target is strongly conserved, just the opposite of PRDM7/9. However this conservation in ZNF133 weakens markedly in the distal 3 repeats. | |||
The 11 conserved zinc fingers in ZNF133 are long enough to specify nearly unique dna sites in a 3 gbp genome, even if not all fingers take part in a given site recognition. Note the SGEKP lockdown cap departs from canonical form in repeats 5, 7, 12, and 13 perhaps impacting binding site utility. Human variation in repeat numbers has not been studied but it appears from phylogenetic considerations to be far less common than in PRDM7/9. [http://www.vivo.colostate.edu/molkit/dnadot/ Dotplots] of ZNF133 show far less agreement across repeats at the dna level, indicating that neither homogenization, expansion, nor contraction of repeats by replication slippage has occurred recently in this gene (unlike PRDM9). | |||
Alignment of human ZNF133 zinc finger array to orthologs in Primates, Glires, Laurasiatheres, Xenarthra and Afrotheres | |||
z z z z z z z z z z z z z z z z z z z z | |||
homSap VNCGECGLSFSKMTNLLSHQRIHSGEKP YVCGVCEKGFSLKKSLARHQKAHSGEKP IVCRECGRGFNRKSTLIIHERTHSGEKP YMCSECGRGFSQKSNLIIHQRTHSGEKP YVCRECGKGFSQKSAVVRHQRTHLEEKT | |||
calJac ............................ ............................ ............................ ............................ ............................ | |||
oryCun ........G...LA.............. ............................ ............................ ...T........................ ............................ | |||
equCab ........G................... ............................ ............................ ............................ ............................ | |||
canFam ...R....G................... ............................ ............................ ...................R........ ............................ | |||
dasNov I..A....G................... ............................ ............................ ............................ .......................S.... | |||
proCap ...E....G................... ............................ ............................ ........R................... .......................S.... | |||
homSap IVCSDCGLGFSDRSNLISHQRTHSGEKP YACKECGRCFRQRTTLVNHQRTHSKEKP YVCGVCGHSFSQNSTLISHRRTHTGEKP YVCGVCGRGFSLKSHLNRHQNIHSGEKP IVCKDCGRGFSQQSNLIRHQRTHSGEKP | |||
calJac ............................ ............................ ............................ ............................ ............................ | |||
oryCun ...G........................ ............................ ............................ .....................T...... ...Q......................R. | |||
equCab ............................ ............................ ............................ .........................D.. ............................ | |||
canFam ...N........................ ............................ ............................ .........................D.. ............................ | |||
dasNov ............................ ............................ ............................ ................I........D.. ............................ | |||
proCap ............................ ...G........................ ............................ ................T........D.. ............................ | |||
> | homSap MVCGECGRGFSQKSNLVAHQRTHSGERP YVCRECGRGFSHQAGLIRHKRKHSREKP YMCRQCGLGFGNKSALITHKRAHSEEKP CVCRECGQGFLQKSHLTLHQMTHTGEKP YVCKTCGRGFSLKSHLSRHRKTTS VHHRLPVQPDPEPCAGQPSDSLYSL | ||
calJac ...A......................K. ............................ ............................ ..........I............N.... ....M..Q..............K. ......L..G...R....A...C.. | |||
oryCun ...Q............L.........K. ............................ .T.........S.........T...... .GGGQ...S.S......S..L..K.... H......Q...Q.........IKA ...KP.LH..S.AYS...PGP.... | |||
2 | equCab ...E............I.........K. ............................ .T........S.........W......L ..........I.....V......Q...L .......Q...Q........RMK. ..Q.P.PH.AS.A.S..S..P.H.. | ||
canFam ...E......................K. ............................ ..........S......I...V...... ........D.I.....L......Q.... ....M.DK...H........RMK. ..YK..LP....A....S..L.H.. | |||
dasNov ...E............I.........K. ...................R........ .T........S...T......L...... ........S.I.R...I....I.KE... ...R...Q...Q.......SRMKC ...KPLL...S.DYS..S..P.... | |||
proCap ...ED...........I.........K. ............................ .A....R...N...T..A..QL...D.L .......ED.M.....LV.....K.... ..SR.H.Q..NQ......Y.RIK. ...KS.F.S.L.T.S..S.VPV... | |||
[[Image:ZNF133function.png|left]] | |||
The ubiquitously expressed ZNF133 has been established by [http://www.e-emm.org/article/article_files/EMM039-04-03.pdf experiment] to be a transcriptional repressor, recognizing specific sites in dsDNA. Despite the presence of the KRAB domain (which usually has this task), the zinc finger array alone contributes to transcriptional repression, with this effect mediated by another gene product, PIAS1, which binds the main array and recruits histone deacylases. The early zinc finger is not necessary for the PIAS1 effect and though conserved, its role remains obscure. PIAS1 may also [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0020321 have a role] in PRDM9 and recombination. | |||
<br clear = all> | |||
[[Image:Znf133Freqs.gif|left]] | |||
For ZNF133, the weblogo below based on 413 repeats from 32 placentals illustrates that quite different selectional pressures have been operative here than in PRDM7/9. First, variation is not concentrated at the four special amino acid positions (purple boxes between CxxC HxxxH) but instead is distributed (though unevenly) among the non-C2H2 positions. Some of this occurs at residues primarily concerned with the zinc binding fold and not targeting macromolecule interactions. This establishes structural variation in the fold can be tolerated, ie PRDM7/9 is the real oddity for not exhibiting it. | |||
<br clear = all> | |||
The early zinc finger (which is [http://pfam.sanger.ac.uk/family?id=zf-C2H2 classified by Pfam] as C2H2)in the terminal exon is rather variable. While a consistently found zinc finger in such a protein is suggestive, nothing can be said about its function at this time. | |||
early zinc finger of ZNF133 early zinc finger of PRDM7/9 early zinc finger of ZNF343 early zinc finger of ZNF589 | |||
homSap YLDPFCPPGFSSQKFPMQHVLCNHPPW HPCPSCCLAFSSQKFLSQHVERNHSSQ YTCSSCLLAFSCQQFLSQHVLQIFLGL YTCSSCLLAFSCQQFLSQHVLQIFLGL | |||
panTro ........................... ........................... ........................... ....C.......P.............. | |||
gorGor ........................... ........................... ..........L................ ........................... | |||
ponAbe ........................... ........................... ........................... ........................... | |||
rheMac ........................... .........................T. .P......................... ........................... | |||
papHam ........................... .........................T. .P......................... ........................... | |||
calJac C.......................... .................H......... .P......................... .......VV.................. | |||
micMur H.G..F..DL......V.R...S.... ......S.............KHT.... .P.......S.........T....Q.. ..FWL...................... | |||
otoGar H.G.L...DL......R...P...... ......S....T..........T.P.. .P..................FR..... (no seqs before duplication) | |||
tupBel H.SVS..LD...E......E....H.. ...L..S.........N....H...C. (no seqs before duplication) | |||
cavPor Q.G..GG.D..A.R..V.....GQ... ......S.....H......M.CS.... | |||
oryCun H.G.L...DC.T..L.V..T..DP... ...FL.S.........T....W..RTE | |||
ochPri S.G.C....L...N....QP.GDP.R. ...A..S.............QH..P.. | |||
turTru H.G..R..D....QLR...M..S.... Q..G..S.......I......CS.P.. | |||
bosTau H.C.....DLC....H..Q...SP... ......S......R........S.P.. | |||
equCab H.C.....D.....VH..R........ .R....S..............CK.... | |||
felCat H.C....SD..-L..H...M..T.... ......S............L.H..P.. | |||
canFam H.C.L..SD.....RHT..M....... ......SV.....T.....GK...P.E | |||
myoLuc H.CA....D......H...M..SN... ......S.................P.. | |||
eriEur PSC.SN..DI....SH...MP...C.. Y...C.S....N.....R...HS.P.L | |||
sorAra H.C....SD.....LHV.R........ ......R............MKHS.P.P | |||
loxAfr QPC.....D......H..R...SP... N.....P..L...QLKHS.PFQSLPGT | |||
proCap Q.C.....N..G...H...A....R.. ......P....TP....H..KHS.PC. | |||
echTel QPC....LD..N...HK.....S.A.. ......P....TE.......Q...P.. | |||
dasNov QFC.....D...K..H......S.... ......P....T.....Y..NHS...E | |||
HKR1 is another zinc finger protein that often surfaces in PRDM9-related blast searches. Structurally it is very similar to ZNF133. The zinc finger array begins with two very degenerate units that cannot bind zinc but may still retain the fold. The next 9 fingers are conventional but the tenth is missing the last two amino acids of the SGEKP cap. The last repeat has an intercalating residue after the two cysteines and lacks the final 3 cap residues. These features were in place at the time of stem placental divergence. | |||
This gene, sometimes called ZNF875, also arose in placental mammal as part of the dramatic expansion of zinc finger proteins. However regulation of gene expression is probably no more refined in placentals than in marsupials, birds and other vertebrates -- these just have different systems. Indeed, rodents seem to have lost both HKR1 and ZNF133 yet get along just fine with poor overall orthologous correlation to primates. | |||
The intra-repeat pattern of variation is different than in PRDM9 and ZNF133. There is more of it and this variation is not concentrated on the macromolecule recognizing amino acid positions, in fact seems to avoid it. This implies that the binding partner is fixed. The single [http://www.ncbi.nlm.nih.gov/pubmed/9813242 1998 publication] on this gene sheds no light on what this might be. Assuming the function is in regulation of gene expression, the recognition sites in human might be predicted approximately from the conserved zinc fingers. This would yield an association with specific genes including false positives and negatives. Repeating this exercise in a dozen mammals and identifying the commonalities to the human gene set would yield a much improved list of regulated genes. HKR1 is widely expressed in a variety of tissues. | |||
z z z z z z z z z z z z z | |||
HKR1_homSap IKYEEFGPGFIKESNLLSLQKTQTGETP YMYTEWGDSFGSMSVLIKNPRTHSGGKP YVCRECGRGFTWKSNLITHQRTHSGEKP YVCKDCGRGFTWKSNLFTHQRTHSGLKP YVCKECGQSFSLKSNLITHQRAHTGEKP | |||
HKR1_panTro2 ............................ ..............I............. .G.......................... ............................ ............................ | |||
HKR1_ponAbe2 ........D.......F.F......... ..............I.........R... ............................ ....H...............GI...... .M.......................... | |||
HKR1_papHam1 ...........................A ..............I............. ............................ ............................ ............................ | |||
HKR1_calJac1 ............K......R.......A .V.....Q.........G.......... R........................... ..........S...........R..... ....D...............K....... | |||
HKR1_tarSyr1 ..C.........N....NF...H....A .......Q..S.V.......K....E.. .M.........................A .........................V.. F........................... | |||
HKR1_micMur1 .......R....DP...GF...H....T .......Q..S..............E.. ...G........................ A.......................V... M.......................... | |||
HKR1_dipOrd1 L...KL..R.M.....P.....HPR..S FIG.K..Q.LSRLP..M...K..V.D.. FL.Q.................M...... F....................I...V.. .M.Q.................S...... | |||
HKR1_equCab2 ...R...L..............H...I. R..S...Q..SN....T..QSMR..E.. ...G............V........... ....E....................VR. .......................S.... | |||
HKR1_canFam2 .......L..L..PK......MGA.... ......KQ..SKR.I....QKIP..EN. ...K........................ ....E....................V.. .......................S.... | |||
HKR1_proCap1 .G.GDL.L...RG.D......AY..G.T .LCN...RDL........KQ..R.R... H..S............L........... H..AE...A.A.R..........A.... HG.RD.....R..A..AA.R...A.AR. | |||
HKR1_dasNov2 ..CTD..F.C..K..V......NIA.SS ...S...EG.N...I....R..Q.EE.. ..........N................. ....E................I...V.. .I.....................S.... | |||
HKR1_choHof1 M.CG...L....K..V......HI...A ...S..ERG.S...I....Q....EE.. ..........N......A.......... ....E................I...V.. .I.....................S.... | |||
z z z z z z z z z z z z z z z z z z z z | |||
HKR1_homSap YVCRECGRGFRQHSHLVRHKRTHSGEKP YICRECEQGFSQKSHLIRHLRTHTGEKP YVCTECGRHFSWKSNLKTHQRTHSGVKP YVCLECGQCFSLKSNLNKHQRSHTGEKP FVCTECGRGFTRKSTLSTHQRTHSGEKP | |||
HKR1_panTro2 ......................................................... ............................ ............................ ................I........... | |||
HKR1_ponAbe2 ............................ ............................ ............................ ............................ ................I........... | |||
HKR1_papHam1 ............................ ............................ ............................ .A.......................... ...A............I........... | |||
HKR1_calJac1 .......H........I..R........ .T.......................... ......W.Q................... ..F......................... ...MA.......R...I........... | |||
HKR1_tarSyr1 ......E.........I..R.I...... .V....K..................... .....................M...... ........R..........R........ ................N........... | |||
HKR1_micMur1 ................I........... .V......A................... ............................ ............................ ................I........... | |||
HKR1_dipOrd1 ...K...S........I........... FV....Q.R.......V........... .I......G.......L........... ........S.......S...KA.A.... .G......S.....S.V...KK.....L | |||
HKR1_equCab2 ................I........... .V.......................... ..........................R. .T......R.................... ..R............I........... | |||
HKR1_canFam2 .......H........I........... .V....D.S................... .I.......................... ........R.................... ..R............I........... | |||
HKR1_proCap1 H..A....A.G.S...A..A......R. HA.GQ...A.G.....V........... F........................I.. ......E................S..... ..Q.........T..V........... | |||
HKR1_dasNov2 F......H....N...I..L........ .V.......................... ...P.....................I.. .M.....................S..... I.R............I........... | |||
HKR1_choHof1 F......H....N...I..L........ .V.......................... .......................A.I.. ................S......S..... K.R...Q........IS.......... | |||
z z z z z z z z z z z z z z z z z z z z | |||
HKR1_homSap FVCAECGRGFNDKSTLISHQRTHSGEKP FMCRECGRRFRQKPNLFRHKRAHSGA FVCRECGQGFCAKLTLIKHQRAHAGGKP HVCRECGQGFSRQSHLIRHQRTHSGEKP YICRKCGRGFSRKSNLIRHQRTHSG | |||
HKR1_panTro2 ............................ .......................... ............................ ............................ ......................... | |||
HKR1_ponAbe2 ............................ .......................... .......................S.... ............................ ......................... | |||
HKR1_papHam1 ...R............V........... .......................... .......................S.... ..........N................. ......................... | |||
HKR1_calJac1 ...R........................ .T........................ ....G...A..D.....N.H.E.S...L ..............Y............. ...........W............. | |||
HKR1_tarSyr1 ...R........................ .......................... ...G.......D....L...KE.SA... ...P....D...K............... .V......C.......V.......R | |||
HKR1_micMur1 ...R........................ .V.......................S C.......S..D...F.......L.... ............................ ........C...........K.... | |||
HKR1_dipOrd1 ...R..K.S....L..HT...I...... .......Q.................D S..........D.......E...S.N.V .M..............L........... ......A.A.......L.....I.. | |||
HKR1_equCab2 ...V........................ .I........G............A.. ...........D.....T.....S...S ................A....I...... .T....................G.. | |||
HKR1_canFam2 ...R.............A.......... .I.....K..S........R.....T ...........D.....T..K..S.... .....................I...... ......E................P. | |||
HKR1_proCap1 ...R....A.....A.L........... ...G......S.R......R.T.... L..K.......D....NA.....S..R. ...V......G.....V........... .V....E.................. | |||
HKR1_dasNov2 ...R........................ .I..Q.....S............... ...........D.....T.....S.... ............................ ......................... | |||
HKR1_choHof1 ...R........................ .I.K......S..............V ....D......D.....T.....S.S.. ......R..................... .V....................... | |||
HRK1 is found within a cluster of ZNF genes on chromosome 19 but has no better than 50% identity to any of them. PRDM7, PRDM9, ZNF133, ZNF343, ZNF589, ZNF169 and ZNF596 are not found in tandem ZNF clusters nor in syntenic associations, as determined by setting the UCSC GeneSorter tool to gene distance and comparing gene neighbors. | |||
[[Image:HKR1cluster.jpg|left]] | |||
<br clear=all> | |||
==== ZNF343 and ZNF589 ==== | |||
ZNF343 is another very closely related KRAB zinc finger array protein. It appears restricted phylogenetically to primates but may have earlier spin-offs reminiscent of PRDM7 (such as the microbat sequence below). Two species, baboon and tarsier have deletions of 2 and 1 zinc finger respectively. The new world monkey Callithrix has a moderately degenerated pseudogene. However it is not plausible that other mammalian species (such as rodents) ever had this gene. | |||
No experimental study has ever considered the function of this gene though occasionally it surfaces in expression studies. It is quite conserved at least in apes, indicative of an important function in gene regulation via specific site recognition. Outside of the terminal zinc finger region, ZNF589 most closely resembles ZNF589 and ZNF133 and so has no direct bearing on PRDM7 or PrDM9. | |||
Prior to ZNF gene family expansion, each of the proteins initially present may have had multiple functions (as proposed by [http://en.wikipedia.org/wiki/Protein_moonlighting Piatigorsky]). With zinc finger arrays, different subsets of fingers may have recognized different dna sites, regulating different genes. After gene duplication, the descendent genes could then diverge to specialize on distinct subsets of these pre-existing functions (subfunctionalization). This allowed fine-tuning of regulation relative to a parent gene operating under slightly conflicting selectional pressures that had to be satisfied simultaneously. | |||
Placental mammals today do not differ greatly from the stem placental mammal in which the expansion began. This expansion (and later contraction) continued into the present era, yielding many lineage-specific sets of ZNF gene families (ie lack of 1:1 orthologous correspondences). Evolution need not take the same path to reach the same end -- indeed, marsupials, birds and other vertebrates have attained excellent regulation of gene expression by other approaches. | |||
ZNF343_homSap INCREYEPDHNLESNFITNPRTLLGKKP YICSDCGRSFKDRSTLIRHHRIHSMEKP YVCSECGRGFSQKSNLSRHQRTHSEEKP YLCRECGQSFRSKSILNRHQWTHSEEKP YVCSECGRGFSEKSSFIRHQRTHSGEKP | |||
ZNF343_panTro ...................S........ ............................ ............................ ................S........... ............................ | |||
ZNF343_gorGor ............................ ............................ ............................ ............................ ............................ | |||
ZNF343_ponAbe .....C...................... ..............A............. ............................ ............................ ............................ | |||
ZNF343_nomLeu .....C...............I...... .....................T...... ............................ ...........N................ ............................ | |||
ZNF343_macMul .....C...RS................. ..............A......T...... ............................ ...........N....K........... ............................ | |||
ZNF343_papHam .....C...RS................. ..............A......T...... ............................ ...........NN...K........... ....D....................... | |||
ZNF343_tarSyr N..K.R...YSP.....R.S..F..E.. CV......G..N....N..R.T..V... ....D.....KNR.T.I.......G... .VR.Q..RG.SQ..NVAQ..R...D... .I.R......RD..TLVI.E........ | |||
ZNF343_otoGar V....F.S.C...........V.FRE.. .V.....PG.....I......T.TG... .E..............T..R........ ...........N....S.......G... .M....E....Q................ | |||
ZNF343_myoLuc GS.N.H.L.CS.K...AV.QV..SEE.. .V.RE...G.NNK.N.N....T...... ...GD......LMAI.VH......G... .V.K...RG.SK..N....TE....... .L.R...QS.RNN.VL....WI...... | |||
ZNF343_homSap YVCLECGRSFCDKSTLRKHQRIHSGEKP YVCRECGRGFSQNSDLIKHQRTHLDEKP YVCRECGRGFCDKSTLIIHERTHSGEKP YVCGECGRGFSRKSLLLVHQRTHSGEKH YVCRECRRGFSQKSNLIRHQRTHSNEKP | |||
ZNF343_panTro ............................ ............................ ............................ ............................ ............................ | |||
ZNF343_gorGor ............................ ............................ ............................ ............................ ............................ | |||
ZNF343_ponAbe ........G................... ...K........................ ...G........................ ............................ ............................ | |||
ZNF343_nomLeu ........G................... ............................ ............................ ............................ ............................ | |||
ZNF343_macMul ........G................... ............................ ............................ ............T............... ............................ | |||
ZNF343_papHam ---------------------------- ---------------------------- .......G.................... ............T............... ............................ | |||
ZNF343_tarSyr F..S.Y.QG.IQ..Q.LV...T....N. ---------------------------- ....K.....SW..H.LV.Q.K...... ...R....S..Q..CVIT.........P .I....G....K..S.........G... | |||
ZNF343_otoGar .I......G.S..........T...D.. ......R.....K.N..R.....SN... .I............N..V...M...... .T.S........................ ......G..Y.............A.... | |||
ZNF343_myoLuc ...P....G.AY.........T...... .I.Q...H...EK.SF.R.....SG... F..L......G.....RK.Q........ .T.S....S.TQ..F..I..G....... ......G.S..Y........K...DV.. | |||
ZNF343_homSap YICRECGRGFCDKSTLIVHERTHSGEKP YVCSECGRGFSRKSLLLVHQRTHSGEKH YVCRECGRGFSHKSNLIRHQRTH | |||
ZNF343_panTro ............................ ............................ ....................... | |||
ZNF343_gorGor ............................ ............................ ..........G............ | |||
ZNF343_ponAbe F........................... ................P........... ....................... | |||
ZNF343_nomLeu ..............S............. ...................K........ ....................... | |||
ZNF343_macMul ............................ .....................I...... ................V...... | |||
ZNF343_papHam ............................ .....................I...... ....................... | |||
ZNF343_tarSyr .V.K......SQ..Y..K.Q...LD... FI.R.......W...............P ...........Q..Y..K.E... | |||
ZNF343_otoGar .V.G..........A............. .IR.DR...S.Q....VS.........W ..........GY........... | |||
ZNF343_myoLuc ..........FY..D..I.......... ........S..Q..F.VI..G....K.. ....D...S..YR....T...K. | |||
ZNF589 is also a primate-specific expansion within the KRAB ZNF gene family which may have expanded independently from the same parental gene in artiodactyls, again similarly to the separate expansions of PRDM7. It has been the subject of 3 publications under the name [http://www.ncbi.nlm.nih.gov/pubmed/12097288,10748030,10029171 SZF1] and its consensus region [http://cancerres.aacrjournals.org/content/62/13/3773.long identified experimentally] as CCAGGGTAACAGCCG which is similar to that of ZBRK1. Regulation of gene expression reportedly takes place in hematopoietic progenitor cells. | |||
In humans, ZNF589 has an internal stop codon at the second cysteine within the 5th repeat due to a T to A transversion. This is not an error or mutation in the reference genome hg19, nor a balanced polymorphism, nor 1% allele as no corrective SNP is known from the 1000 genome project or individual sequencing projects. It remains possible that some human populations (notably African because of greater diversity) will not have this stop codon if it is a very recent development. However all human populations sampled including bushman KB1 genomic reads all contain it, as do Neanderthal and Denisova fossil dna. (View chr3:48,285,273-48,285,286 on [http://genome-test.cse.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr3:48285273-48285286 UCSC human genome browser] hg19 with appropriate tracks opened.) | |||
Past the stop codon, the zinc finger array continues on another nine repeats and do not seem impaired (strict conservation of cysteines, histidines, and invariant phenylalanine and leucine). It is not clear whether the mRNA would be targeted by [http://en.wikipedia.org/wiki/Nonsense-mediated_decay nonsense mediated decay], whether a truncated and possibly still functional protein is produced, or whether that a suppressor mechanism that allows some read-through of the early stop codon. If ZNF589 functions with four repeats, the dna recognition sites would be truncated relative to what the full zinc finger array could have recognized. Terminal alternative splicing has been seen for ZNF589 but not rejoin the array. | |||
Chimpanzee does not have this internal stop codon and has a full set of repeats. However the orthologous gene in gorilla (contig CABD02243014) has a frameshift near the end of its 10th repeat. Assuming this is not assembly error in a low coverage genome, this raises the same question about pseudogenization vs function-retaining truncation as in human. If this represents gene loss, the event is independent from the one in human and the resultant protein may have higher dna specificity (recognize a longer or different site). | |||
Similarly orangutan has a frameshift at the end of the 7th repeat, a reading frame restoring frameshift 4 repeats later followed shortly by an early stop codon. Gibbon also has a frameshift. However macaque, like chimpanzee, has maintained a full length zinc finger array. New world monkey Callithrix has a much older pseudogene. This is truly an ortholog because biflanking synteny is still preserved (NME6+ ZNF589- CAMP-) from human to marmoset. | |||
Tarsier also has severely decayed gene candidate. Its contig maps uniquely to the ZNF589 region of human by blastn but is too short to establish flanking synteny. Since ZNF589 appears missing in lemurs and tree shrew, the duplication event occurred in the tarsier divergence stem, or about 65 myr ago. Tarsiers are basal haplorrhine and [http://www.ncbi.nlm.nih.gov/pubmed/21620437 not part of the strepsirrhine (lemur) clade]. | |||
Thus the gene duplicate ZNF589 persisted in the ancestor past the tarsier and new world monkey divergences and losses, kept a full length repeat role in old world monkeys and chimp, but may have carved out altered separate roles in great apes with utilizing recently truncated arrays. | |||
The parental gene for ZNF589 -- based on the terminal exon minus the array -- appears to be ZNF133 or PRDM7, genes that arose about the same time in stem placentals. | |||
ZNF589 Repeat Region in Apes: | |||
nominal length of zinc finger array produced shown in <span style="color: #0066CC;">blue</span> | |||
stop codons and truncated repeats shown in <span style="color: #FF0000;">red</span> | |||
frameshifts and cryptic repeats shown in <span style="color: #990099;">purple</span> | |||
<table><tr align="left" valign="top"> | |||
<td> | |||
<span style="color: #0066CC;">>ZNF589_homSap AADD01032563 | |||
QVCRECGRGFSRKSQLIIHQRTHTGEKP | |||
YVCGECGRGFIVESVLRNHLSTHSGEKP | |||
YVCSHCGRGFSCKPYLIRHQRTHTREKS | |||
FMCTVCGRGFREKSELIKHQRIHTGDKP | |||
YVCRD</span><span style="color: #FF0000;">*GRGFVRRSCLNTHQRIHSDEKP | |||
FVCRECGRGFRAKSTLLLHQWTHSEVKP | |||
HVCEECGHGFSQKSSLKSHRRTHSGEKP | |||
YVCGECGRGFSRRIVLNGHWRTHTGEKP | |||
YTCFECGRNFSLKSALSVHQRIHSGEKP | |||
YACTECGQGFITKSQLIRHQRTHTGEKP | |||
YVCGECGRGFIAQSTLHYHRSTHSKEKP | |||
YVCSQCGRGFCDKSTLLAHEQTHSGEKP | |||
YVCGECGRGFGRKILLNRHWRTHTGEKP | |||
YACIECGRNFSHKSTLSLHQRIHSGEKP | |||
YACVECGQSFRRKSQLIIHQKIHSGKSF | |||
RGARSEDVILATSQPSATPAEMLREKPCL | |||
</span></td> | |||
<td> | |||
<span style="color: #0066CC;">>ZNF589_panTro AADA01029841 | |||
QVCRECGRGFSRKSQLIIHQRTHTGEKP | |||
YVCGECGRGFIVESVLRNHLSTHSGEKP | |||
YVCSHCGRGFSCKPYLIRHQRTHTREKS | |||
FMCTVCGRGFREKSELIKHQRIHTGDKP | |||
YVCRDCGRGFVRRSCLNAHQRIHSDEKP | |||
FVCRECGRGFRAKSTLLLHQWTHSEVKP | |||
HVCEECGHGFSQKSSLKSHRRTHSGEKP | |||
YVCGECGRGFSRRIVLNGHWRTHTGEKP | |||
YTCFECGRNFSLKSALSVHQRIHSGEKP | |||
YACTECGQGFITKSQLIRHQRTHTGEKP | |||
YVCGECGRGFIAQSTLHYHRSTHSKEKP | |||
YVCSQCGRGFCDKSTLLAHEQTHSGEKP | |||
YVCGECGRGFGRKILLNRHWRTHTGEKP | |||
YACIECGRNFSHKSTLSLHQRIHSGEKP | |||
YACVECGRSFRRKSQLIIHQKIHSGKSF | |||
RGARSEDVILATSQPSATPAEMLREKPCL | |||
</span></td> | |||
<td> | |||
<span style="color: #0066CC;">>ZNF589_gorGor CABD02243014 | |||
QVCRDCGRGFSRKSQLIIHQRTHTGEKP | |||
YVCGECGRGFIVESVLRNHLSTHSGEKP | |||
YVCSHCGRGFSCKPYLIRHQRTHTREKS | |||
FMCTVCGRGFREKSELIKHQRIHTGDKP | |||
YVCRDCGRGFVRRSCLNTHQRIHSDEKP | |||
FVCRECGRGFRAKSTLLLHQWTHSEVKP | |||
HVCEECGHGFSQKSSLKSHRRTHSGEKP | |||
YVCGECGRGFSRRIVLNGHWRTHTGEKP | |||
YTCFECGRNFSLKSALSVHQRIHSGEKP | |||
YACTECGQGFITKSQLIRHQRTHT</span><span style="color: #990099;">gEKP | |||
YVCGECGRGFIAQSTLHYHRSTHSKEKP | |||
YVCSQCGRGFCDKSTLLAHERTHSGEKP | |||
YVCGECGRGFGRKILLNRHWRTHTGEKP | |||
YACIECGRNFSHKSTLSLHQRIHSGEKP | |||
YACMECGRGFRRKSQLIIHQKIHSGKSF | |||
RGARSEDVILATSQPSATPAEMLREKTCL | |||
</span></td> | |||
<td> | |||
<span style="color: #0066CC;">>ZNF589_ponAbe ABGA01071880 | |||
QVCRECGRGFSRKSQLIIHQRTHTGEKP | |||
YVCRECGRGFIVESVLRNHLSTHSGEKP | |||
YVCSHCGRGFSCKPYLIRHQRTHTREKS | |||
FMCTVCGQGFREKSELIKHQRIHTGDKP | |||
YVCRDCGRGFVRRSCLNTHQRIHSDEKP | |||
FVCKECGRGFHAKSTLLLHQWTHSEVKP | |||
HVCEECGHGFSQKSTLKSHRRTHSG</span><span style="color: #990099;">e</span><span style="color: #990099;">KS | |||
YVCEECGRGFSRRIFLNGHWRTHTREKP | |||
YTCFECGRNFSLKSALSVHQRMHSGEKP | |||
YACTECGQGFITKSQLIRHQRTHTGEKP | |||
YVCREWARLYSSDNPPLPPAYTLQGETp</span> | |||
<span style="color: #0066CC;">YVCSQRG</span><span style="color: #FF0000;">*GFCDKSTLLAHEQTHSGEKP | |||
YVCGECGWGFGRKILLNRHWRTHTGEKT | |||
YACIECGQNFSHKSTLSLHQRIHSGEKP | |||
YACMECGRGFRRKSQLIIHQKIHSGKSF | |||
RGASSEDVILATSQPSATPAEMLREKTCL | |||
</span></td> | |||
<td> | |||
<span style="color: #0066CC;">>ZNF589_nomLeu ADFV01172942 | |||
QVCRECGRGFSRKSQLIIQQRTHTGEK | |||
YVCEECGRGFIVESVLRNHLSAHSAEKP | |||
YVCSHCGRGFSCKPYLMRHQRTHTREKS | |||
FMCTVCGRGFREKSELIKHQIIHTGGKP | |||
YVCRDCGRGFVRRSCLNTHQRIHSDEKP | |||
FVCRECGRGFRAKSTLLLHQWTHSEVKP | |||
HVCEDCGHGFSQKSTLKSHRRTHSGEKP | |||
YVCGECGQGFSRRIFLNGHWRTYTGEKP | |||
YTCFECGRNFSLKSALSVHQRIYWGE</span><span style="color: #990099;">kP | |||
YACVECGRGFITKSQLIRHQRTHTGEKP | |||
YVCGECGQGFIAQSALRYHRSTHSREKP | |||
YVCSQCGrGEAFVINQLAHEQTHSGEKP | |||
YVCGECGQGFGRKILLNRHWRTHTGEKP | |||
YACIECGRNFSHKSTLSLHQRIHSGEKP | |||
YACTECGRGFRRKSQLITHQKTHSGKSF | |||
RGARSEDVILATSQPSATLAEMLREKACL | |||
</span></td></tr> | |||
<tr align="left" valign="top"> | |||
<td> | |||
<span style="color: #0066CC;">>ZNF589_rheMac AANU01238696 | |||
QVCGECGRGFSRKSQLIIHQRTHTGEKP | |||
YVCGECGRGFIVESVLRNHLSTHSGEKP | |||
YVCSQCGRGFSCKPYLIRHQRTHTREKS | |||
FMCTVCGRGFREKSELIKHQRIHTGDKP | |||
YVCRDCGRGFVRRSCLNTHQRIHSDEKP | |||
FVCRECGRGFRAKSTLLLHQWTHSEVKP | |||
HVCEECGHGFSQKSTLKSHQRTHSGEKP | |||
YVCGECGRGFSRRIFLSGHWRTHTGEKP | |||
YTCFECGRNFSLKSALSVHQRIHSGEKP | |||
YACAECGRGFITKSQLIRHQRTHTGEKP | |||
YVCGECGRGFIAQSTLHYHRSTHSGEKP | |||
YVCSQCGRGFRDKSALLAHEQTHSGEKP | |||
YVCGECGWGFGRKILLSRHWRTHTGEKP | |||
YACMECGRNFSHKSTLSLHQRIHSGEKP | |||
YACTECGRGFRRKSQLSIHQKTHLGKSF | |||
RGARSEDVIFASQPSAAPAEMLREKPCL</span> | |||
</td> | |||
<td> | |||
<span style="color: #0066CC;">>ZNF589_papHam ti|2005908815 | |||
QVCRECGRGFSRKSQLIIHQRTHTGEKP | |||
YVCGECGRGFIVESVLRNHLSTHSGEKP | |||
YVCSQCGRGFICKPYLIRHQRTHTREKS | |||
FMCTVCGRGFRENSELIKHQRIHTGDKP | |||
YVCRDCGRGFVRRSYLNTHQRIHSDEKP | |||
FVCRECGRGFRAKSTLLLHQWTHSEVKP | |||
HVCEECGHGFSQKSTLKSHRRTHSGEKP | |||
YVCGECGRGFSRRIFLSGHWRTHTGEKP | |||
YTCFECGRNFSLKSALSVHQRIHSGEKP | |||
YACAECGRGFITKSQLIRHQRTHTGEKP | |||
YVCGECGRGFIAQSTLHYHRSTHSGEKP | |||
YVCSQCGRGFRDKSTLLAHEQTHSGEKP | |||
YVCGECGRGFGRKILLSRHWRTHTGEKP | |||
YACMECGRNFSHKSTLSLHQRIHSGEKP | |||
YACTECGRGFRRKSQLNIHQKTHLGKSF | |||
RGARSEDVIFASQPSAAPAEMLREKPCL</span> | |||
</td> | |||
<td> | |||
<span style="color: #0066CC;">>ZNF589_calJac ACFV01038884 | |||
QMCTVCGRGIRNKSHLIQHQRIHTGDKP | |||
YVCRNCGRGFVRSCLIK HQRILSGEKP | |||
FICRECGRGFRDKSTPHT</span><span style="color: #990099;">hQRAHSGEKP | |||
s</span><span style="color: #0066CC;">CGEECGRGFTRKSTLKSHRRTHSGEKP | |||
YVYGECGWGFSSKGVLNTHWRTHTGAKP | |||
YACRVATSPL</span><span style="color: #990099;">sHKSTLSSHQRIHSGEKP</span> | |||
</td> | |||
<td> | |||
<span style="color: #0066CC;">>ZNF589_tarSyr ABRT010411760 | |||
SVCREYGQSFSRKSHLLRHWRTHTGEKP | |||
YV</span><span style="color: #990099;">nGNCGHTFIDKSVLHNYQSTHSGKKP | |||
YVCRECGCSLD<span style="color: #FF0000;">*</span>KSHLIRHQRTHTQERP | |||
FMCTV</span> | |||
</td> | |||
<td></tr></table> | |||
By comparing each sequence to itself and to PRDM9, it emerges that PRDM9 is highly unusual for its remarkable self-similarity in its zinc finger array. That strongly suggests some form of homogenization (master-slave) of repeats that is unique to it and very likely highly relevant to its role in defining recombination hotspots. Whatever the precise evolutionary relationships to the other closely related zinc fingers, that has not resulted in retention of close matching at the dna level to PRDM9. | |||
[[Image:DotPlotCompZNF.gif|left]] | |||
<br clear=all> | |||
The alignment below of the final exon (minus the terminal arrays) shows positions conserved at 60% identity. Overall, conservation is unremarkable outside the early zinc finger region. However several distal patches also suggest a moderate level of conservation pressure and thus some function beyond merely serving as a long linker between the terminal array (which is wrapped around the major groove of the dna target) and the early repeat (whose binding partner, if any, is unknown). | |||
first anomalous repeat | |||
PRDM9_homSap E.KPEI..CPSC.LAFSSQ.FLSQHV..NH..Q.F....A...L.P.NP.PGDQ.Q.-QQ..D.........GQE....S..L..RT..R....AFSSPP.-.Q..S.R.G.R..E.E....Q..NP..T.K............ ....ECG.GFS..S....HQRTHTGEK. | |||
PRDM7_homSap E.KPEI..CPSC.LAFSSQ.FLSQHV..NH..Q.F....A...L.P.NP.PGDQ.Q.-.Q..D.........GQE....S..L..RT..R....AFSSPP.-.Q..SSR.G.R..E.E....Q..NP..T.K............ ....ECG.GFS..S....HQRTHTG.KP | |||
PRDM7_calJac E.KPEI..CPSC.LAFSSQ.FLS.HV..NH..Q.F........L.P.NP.PG.Q...-QQ..D.........GQE....S..L..RT..R....AFS.PP.-.Q..SSR...R..E.E....Q..NP..T.K............ ....ECG.GFS..S....HQRTHTGEKP | |||
ZNF133_homSap ...PE....P.C...FSSQ.F..QHV..NH....F....A.....P..P.PGDQ...-QQ............G.E.....-.L..RT..R-...AFS.PP.-.Q..SSR.G.R..E.E....Q..NP..T.K............ ....ECG..FS.......HQR.H.GEKP | |||
ZNF589_homSap E.KPE...CPSC.LAF.SQ.FLSQ....NH....F.---A...L.P.NP.P.DQ.Q.-Q...D..........Q........L...T..R.....FSS...-....SS..G.R..E......Q..................... ....ECG.GFS..S....HQRTHTGEK | |||
ZNF343_homSap E.KPEI..C.SC.LAFS.Q.FLSQHV.-----Q.F....A.....P.N..PG...Q..QQ............GQE....S.....RT..R....AF.SP..-.Q..S.R.G....E.E....Q..NP....K............ ....E........S......RT..G.KP | |||
HKR1_homSap E.KPEI...PSC.L.FSSQ..LSQHV...H..Q.F....A...L......P.DQ.Q.----.D................S..L..R........A.SSPP...Q...S.....................T.K............ ....E.G.GF...S.....Q.T.TGE.P | |||
== Domain by domain structure/function == | |||
PRDM7 and PRDM9 are chimeric proteins comprised of 6 recognizable domains joined by linker regions. While multi-domain proteins are common in the overall human proteome, this particular combination occurs nowhere else. However some of the domains here occur in other combinations in other proteins, notable in the vast heterogeneous family of zinc finger proteins (gene names ZNFxxx). | |||
KRAB_A Kruppel | |||
SSXRD | |||
zinc knuckle | |||
PR or SET domain | |||
early zinc finger | |||
terminal zinc finger array | |||
Because the inter-domain linkers are evolving chaotically in terms of little amino acid property conservation and sometimes length, they cannot plausibly be under significant selective pressure, nor can they assume a stable structural fold. However this does not imply that the domains that they link do not have significant physical interactions important to the global tertiary protein structure. To date, only the isolated domains have been studied crystallographically (with the exception of the knuckle-PR combination). | |||
While the domain folds individually are quite ancient and do not reflect de novo innovation in vertebrates from random dna strings, their assembly into PRDM7/9 is fairly recent, about 150 million years ago. Prior to this, a proto-PRDM7 containing the last 4 domains arose and persisted for 300 million years, giving rise to several gene duplicates, all with vaguely understood function related to transcriptional regulation. | |||
The following sections consider what is known about each domain in turn primarily from the perspective of comparative genomics. As of July 2011, 51 land vertebrate genomes are available, providing a rich history of how PRDM7 has been evolving in various branches of the phylogenetic tree. | |||
=== Reciprocal translocation: origin of the SSX1-PRDM chimera === | |||
Upon blastp of the first 6 exons of any PRDM7/9 protein against GenBank restricted to human, SSX1 emerges as the only full length non-self match. Comparison of its 6 exons establishes further that their intron phasing is an exact match. Since this is impossibly coincidental, it follows that PRDM7 (the immediate parent of PRDM9 in primates) arose as a chimera of ancestors to these two proteins prior to marsupial divergence. The percent identity has dropped from the initial perfect agreement to 32% today, without however loss of KRAB_A and SSXRD domain recognizability in either gene family. No other proteins in the human genome -- in particular no zinc finger proteins -- contain these 6 exons though the KRAB domain alone is widespread. | |||
>SSX1_homSap >PRDM9_homSap | |||
<span style="color: #0066CC;">0 MNGDDTFAKRPRDDAKASEKRSK 0 0 MSPEKSQEESPEEDTERTERKPM 0</span> | |||
<span style="color: #990099;">0 AFDDIATYFSKKEWKKMKYSEKISYVYMKRNYKAMTKL 1 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1</span> | |||
<span style="color: #0066CC;">2 GFKVTLPPFMCNKQATDFQGNDFDNDHNRRIQ 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1</span> | |||
<span style="color: #990099;">2 VEHPQMTFGRLHRIIPK 0 2 VKPPWMALRVEQRKHQK 0</span> | |||
<span style="color: #0066CC;">0 IMPKKPAEDENDSKGVSEASGPQNDGKQLHPPGKANISEKINKRS 1 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1</span> | |||
<span style="color: #990099;">2 GPKRGKHAWTHRLRERKQLVIYEEISDPEEDDE* 2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1</span> | |||
PRDM9 <span style="color: #0066CC;">MSPEKSQEESPEEDTERTERKPM</span><span style="color: #990099;">VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI</span><span style="color: #0066CC;">GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ</span> | |||
<span style="color: #0066CC;">M+ + + + P +D + +E++ </span><span style="color: #990099;"> K AF DI+ YF+K+EW +M EK Y +KRNY A+ +</span><span style="color: #0066CC;">G + T P FMC+ +QA Q +D D + R Q</span> | |||
SSX1 <span style="color: #0066CC;">MNGDDTFAKRPRDDAKASEKRSK</span><span style="color: #990099;">---AFDDIATYFSKKEWKKMKYSEKISYVYMKRNYKAMTKL</span><span style="color: #0066CC;">GFKVTLPPFMCN-KQATDFQGNDF---DNDHNRRIQ</span> | |||
> | PRDM9 <span style="color: #990099;">VKPPWMALRVEQRKHQK</span><span style="color: #0066CC;">GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL</span><span style="color: #990099;">ELRKKETERKMYSLRERKGHA-YKEVSEPQDDDYL 1</span> | ||
<span style="color: #990099;">V+ P M R K</span><span style="color: #0066CC;"> MPK +E+ K +S ASG + K + P G+A+ S + ++ </span><span style="color: #990099;"> ++ + LRERK Y+E+S+P++DD </span> | |||
SSX1 <span style="color: #990099;">VEHPQMTFGRLHRIIPK</span><span style="color: #0066CC;">IMPKKPAEDENDSKGVSE------ASGPQNDGKQLHPPGKANISEKINK-RS</span><span style="color: #990099;">GPKRGKHAW-THRLRERKQLVIYEEISDPEEDDE* </span> | |||
This chimera arose subsequent to the duplication of proto-PRMD7 and its divergence to PRDM11, its nearest PRDM relative which has leading exons unrelated to SSX1. Indeed none of the other 14 PRDM proteins have a KRAB or SSXRD domain. The SSX1 gene itself, then and now, lies in a tandem array and so did not disappear as a standalone gene family as only one copy was used up in forming the hybrid protein. For viability, the event was likely a reciprocal translocation, accounting for the SSX array and PRDM7 being on different chromosomes today. | |||
The SSX1-group genes occurs in the human reference genome as 11 features in two nearby clusters both on chromosome X. Some of these may be pseudogenes. The degree of similarity suggests recent gene duplication and/or gene conversion. The array is notorious for reciprocal translocations involving the one of 24 human synaptotagmins, the SYT4 gene on chromosome 18. These translocations [http://www.ncbi.nlm.nih.gov/pubmed/11368913 fuse] early exons of SYT4 with distal exons of an SSX gene, usually SSX1 or SSX2 but sometimes SSX4. The event takes place within intron 4 of the SSX genes and preserves reading frame, allowing for a chimeric protein with disastrous regulatory properties to emerge -- nearly all cases of synovial sarcomas arise from repeated occurrence of this event. | |||
SSX1b + chrX:47967088-47980069 similar to SSX1 | |||
SSX5 - chrX:48045656-48056199 synovial sarcoma X breakpoint 5 | |||
2 | SSX1a + chrX:48114797-48126879 synovial sarcoma X breakpoint 1 | ||
SSX9 - chrX:48154885-48165614 synovial sarcoma X breakpoint 9 | |||
SSX3 - chrX:48205863-48216142 synovial sarcoma X breakpoint 3 | |||
SSX4 + chrX:48242968-48252785 synovial sarcoma X breakpoint 4 | |||
SSX4B - chrX:48261524-48271344 synovial sarcoma X breakpoint 4B | |||
SSX8 + chrX:52651985-52662998 similar to SSX8 | |||
SSX7 - chrX:52673111-52683950 synovial sarcoma X breakpoint 7 | |||
SSX2a - chrX:52725946-52736249 synovial sarcoma X breakpoint 2 | |||
SSX2b + chrX:52780308-52790617 synovial sarcoma X breakpoint 2 | |||
Possibly the SSX1 array has long been predisposed to translocation events. It might seem very difficult to establish the structure of the ancestral array at the time of PRDM chimera formation -- contemporary marsupial has barely related genes on different chromosomes; elephant and dog too lack a multi-gene array. However rhesus but not marmoset has a chr X cluster, so that aspect is restricted to old world primates. A single SSX1 gene can be recovered from elephant but is already quite diverged from human. Marsupials have no evident SSX1 genes today. | |||
This gene fusion of SSX1 and PRDM brought together a negative regulatory domain for transcription with a histone methylase and dna site recognition domain. This new combination succeeded in replacing whatever prior mechanism existed for meiotic breakpoint pairing and recombination. | |||
>SSX1_loxAfr | |||
0 VNRDSSLAKSSKEDTQKPEKESK 0 | |||
2 | 0 AFKDILKYFSKEEWAKLGYSKKVTYVYMKRNYDTMTNL 1 | ||
2 | 2 GLRATLPPFMDPNRLATKSQLDESDEEQNPGTQ 1 | ||
2 DEPPQMASSVRESKHLM 0 | |||
2 | 0 MKPKKPSKEENGSKVVPGTAGLMRTSGPEQAQKQPCPPGKANTSGQQSKQTP 1 | ||
2 | 2 VPGKEETKVWACRLRERKNLVAYEEISDPEEED* | ||
1 | === The zinc knuckle preceding the PR (SET) domain === | ||
2 | |||
A 2011 [http://www.ncbi.nlm.nih.gov/pubmed/21604305 crystallographic study] establishes that a short motif YC..C..........C..HGP found in 6 members of the human PRDM gene family binds zinc via the 3 cysteines and a histidine. The fold most closely resembles the previously known RanBP2 zinc finger domain which occurs in some 21 human proteins, notably nucleoporins NUP153, NUP358, NPL4, EWS, TLS, RBP56, RBM5, RBM10, TEX13A, RANDB2 and ZRANB2. Not all these domains are necessarily homologous because the fold is small and zinc fingers seem to have evolved numerous times. Such fingers can bind other proteins, ssRNA and likely DNA. Their function in PRDM genes is completely unknown but the aromatic residue preceding the first cysteine may contribute to a pi-bonding base stack with guanines. | |||
[[Image:KnuckleSET.jpg|left]] | |||
The domain begins at a phase 2 exon, meaning that the first codon letter is borrowed from the preceding exon splice donor. A dozen earlier residues from this exon are also used but do not exhibit any conservation outside their orthology class. In most cases the knuckle domain exon also contains a downstream PR(SET) domain but at variable intervening lengths (distances shown are to conserved FGP in center of PR(SET) domain. The function of these intervening residues are unknown. | |||
<br clear=all> | |||
exon 6 splice exon 7 SET gene name | |||
IPLNQHTSDPNN 1 2 R<span style="color: #FF0000;">C</span>DM<span style="color: #FF0000;">C</span>ADNRNGE<span style="color: #FF0000;">C</span>PM<span style="color: #FF0000;">HGP</span>LHSLRRLVG .49. PRDM6_homSap | |||
PDPPRPFDPHDL 1 2 W<span style="color: #FF0000;">C</span>EE<span style="color: #FF0000;">C</span>NNAHASV<span style="color: #FF0000;">C</span>PK<span style="color: #FF0000;">HGP</span>LHPIPNRPV .16. PRDM10_homSap | |||
MAEDGSEEIMFI 1 2 W<span style="color: #FF0000;">C</span>ED<span style="color: #FF0000;">C</span>SQYHDSE<span style="color: #FF0000;">C</span>PE<span style="color: #0066CC;">L</span><span style="color: #FF0000;">GP</span>VVMVKDSFV .99. PRDM15_homSap | |||
GSKENMATLFTI 1 2 W<span style="color: #FF0000;">C</span>TL<span style="color: #FF0000;">C</span>DRAYPSD<span style="color: #FF0000;">C</span>PE<span style="color: #FF0000;">HGP</span>VTFVPDTPI .36. PRDM4_homSap | |||
IVPKSFQQVDFW 1 2 F<span style="color: #FF0000;">C</span>ES<span style="color: #FF0000;">C</span>QEYFVDE<span style="color: #FF0000;">C</span>PN<span style="color: #FF0000;">HGP</span>PVFVSDTPV .42. PRDM11_homSap | |||
KEVSEPQDDDYL 1 2 Y<span style="color: #FF0000;">C</span>EM<span style="color: #FF0000;">C</span>QNFFIDS<span style="color: #FF0000;">C</span>AA<span style="color: #FF0000;">HGP</span>PTFVKDSAV .42. PRDM9_homSap | |||
KEISEPQDDDYL 1 2 Y<span style="color: #FF0000;">C</span>EM<span style="color: #FF0000;">C</span>QNFFIDS<span style="color: #FF0000;">C</span>AA<span style="color: #FF0000;">HGP</span>PTFVKDSAV .42. PRDM7_homSap | |||
QEIWDPQDDDYL 1 2 Y<span style="color: #FF0000;">C</span>EE<span style="color: #FF0000;">C</span>QTFFLET<span style="color: #FF0000;">C</span>AV<span style="color: #FF0000;">HGP</span>PKFVQDSVM .42. PRDM7_monDom | |||
NENYRPEDDDYL 1 2 Y<span style="color: #FF0000;">C</span>EI<span style="color: #FF0000;">C</span>QTFFLEK<span style="color: #FF0000;">C</span>VL<span style="color: #FF0000;">HGP</span>PVFVQDLPV .42. PRDM7_ornAna | |||
EEQDDTFNDQPF 1 2 Y<span style="color: #FF0000;">C</span>EM<span style="color: #FF0000;">C</span>QQHFIDQ<span style="color: #FF0000;">C</span>ET<span style="color: #FF0000;">HGP</span>PSFTCDSPA .42. PRDM7_danRer | |||
TEEEELRDEEYF 1 2 F<span style="color: #FF0000;">C</span>EE<span style="color: #FF0000;">C</span>KSFFIEE<span style="color: #FF0000;">C</span>EL<span style="color: #FF0000;">HGP</span>PLFIPDTPA .42. PRDM7_salSal | |||
IKEEEADVKDFL 1 2 Y<span style="color: #FF0000;">C</span>EV<span style="color: #FF0000;">C</span>KSVFFSK<span style="color: #FF0000;">C</span>EV<span style="color: #FF0000;">HGP</span>ALFIADSPV .42. PRDM7_ictPun | |||
YV<span style="color: #FF0000;">C</span>RE<span style="color: #FF0000;">C</span>GRGFSWQSVLLT<span style="color: #FF0000;">H</span>QRT<span style="color: #FF0000;">H</span>TGEKP comparison to longer zinc finger in main array of PRDM7/9 | |||
=== Structural alignment of all PRDM proteins === | |||
To determine the evolutionary relationship of the 16 human PRDM genes, it is useful (given the great divergence in primary sequence) to consider rare genomic events such as intron gain/loss and indels. Only 7 of the 16 contain the knuckle region. Of these PDRM11 is the most closely related to PRMD9. | |||
This is fortunate because the 3D structure of PRDM11 was recently determined (PDB: 3RAY) from before the knuckle region on into the final exon, thus allowing threading of PRDM9 (whose structure has not been studied). The dozen-odd conserved patches in these widely diverged paralogs find their explanation in the atomic details of this structure. Note the PRA(SET) domain and zinc fingers are all that can currently modeled as the KRAB, SSXRD and final exon have no counterparts at PDB. | |||
The knuckle region apparently represents a one-time domain acquisition relative to a knuckle-less ancestral state. The date of this event relative to species phylogeny and the source of the domain are unclear (it is very unlikely to have evolved in situ). Similarly, the internal phase 00 intron is ancestral even though it breaks up a coherent structural domain. Note the final 12 intron is also ancestral -- the PR(SET) domain never occurs without it even though zinc fingers are not always found in the next exon. However the later 21 intron is a newer acquired feature specific to PRDM9 and its closest associates, post-dating acquisition of the knuckle domain and predating duplication and divergence of the PRDM7/9 group. This again follows from gene tree and parsimony considerations. | |||
Crystallographic coverage is excessive yet highly unsatisfactory -- the knuckle-PR(SET) domain is covered by 6 different structures, yet none of them are exactly what is needed (PRDM11 3RAY; PRDM4 2L9Z/3DB5; PRDM10 3IXH; PRDM1 3DAL; PRDM2 3JV0; PRDM12 3EP0). There is no coverage of the preceding KRAB or SSXRD domain or the following early knuckle. However on the knuckle-PR(SET) domain, all these structures could likely be superimposed simultaneously on the near-universal domains identified below. PRDM7/9 would then follow this fold trace as well, though it could be modeled directly from just PRDM11. The intervening regions between conserved anchors can be modeled for PRDM7/9 only to the extent that local conservation in length and residue can be found to a determined structure. For example, <span style="color: #FF0000;">IFY</span>RTCRV<span style="color: #FF0000;">I</span> in PRDM9 can be modeled by the PRDM11 structure since its internal residues contain three matches and no gaps, <span style="color: #FF0000;">IFY</span><span style="color: #0066CC;">R</span>A<span style="color: #0066CC;">CR</span>D<span style="color: #FF0000;">I</span>. | |||
Humans have [http://www.ncbi.nlm.nih.gov/pubmed/21564555 51 genes encoding SET domains], with the PRDM group most diverged from the canonical structures. It is difficult enough to meaningfully align the PRDM and even more so to include all 51 of these lysine methylases. [http://onlinelibrary.wiley.com/store/10.1111/j.1747-0285.2011.01135.x/asset/supinfo/CBDD_1135_sm_TableS1-FigS1-4.pdf?v=1&s=81ef430033f1fd8d18bed72183e312564387ad66 When that is done], most of the conserved patches below emerge as universal motifs yet others are restricted to the PRDM family. All of these proteins would likely bind S-adenosyl methionine and have a lysine pocket in addition superimposable global folds (neither relatable to the 45 human arginine methyltransferases). | |||
[[Image:PRDMs.gif|left]] | |||
0 | <br clear=all> | ||
gapping: uncertain between conserved markers iM: initial methionine, protein thus too short for further comparison | |||
2 | knuckle: shortened zinc finger motif underlining: magenta coloring shows non-informative idiosyncratic introns | ||
C2H2: terminal zinc finger region following universal phase 12 intron PRDM15: duplicated diverged exon removed 21 SWPASGHVHTQAGQGMRGYEDRDRADPQQLPEAVPAGLVRRLSGQQLPCRSTLTWGRLCHLVAQGR | |||
0: indel unifying PRDM9/7/11, cannot be resolved as insertion or deletion 7: near-universal motif NWMrYV split by phase 21 intron gained by PRDM9/7/11/4 | |||
1: arginine supporting PRDM6 as outgroup to the knuckle subgroup 8: inexplicable repositioning of 6 residues to previous exon in PRDM4 | |||
2: near-universal motif SLP 9: near-universal motif EQNL | |||
3: near-universal motif GF 10: near-universal motif IFY | |||
4: indel unifying PRDM9/7/11, resolvable as an insertion 11: near-universal motif ELLVWY | |||
5: near-universal motif FGP 12: possible synapormorphy grouping first 9 genes | |||
6: near-universal motif WLI split by universal phase 00 intron PRDM16: CVDANQAGAG insertion removed from ISEDLGSEKFCVDANQAGAGSWLKYIRVA | |||
PRDM3: inexplicably has official gene name MECOM text-pdf version [http://genomewiki.ucsc.edu/index.php/Image:PRDMseq.pdf here] | |||
Applicable 3D structural determinations: | |||
> | .... PRDM9 Y<span style="color: #FF0000;">C</span>EM<span style="color: #FF0000;">C</span><span style="color: #990099;">QNFFIDS</span><span style="color: #FF0000;">C</span>AA<span style="color: #FF0000;">H</span>GPPTFVKDSAVDKGHPN<span style="color: #FF0000;">R</span>SAL<span style="color: #FF0000;">SLP</span>PGLRIGPSGI.PQAGL<span style="color: #FF0000;">GV</span>WNEASDLPLGLH<span style="color: #FF0000;">FGP</span>YEGRIT.....EDEEAANNGYS<span style="color: #FF0000;">WLI</span>TKG.RNCYEY<span style="color: #FF0000;">VD</span>.......GKDKSWA<span style="color: #FF0000;">NWM</span>R<span style="color: #FF0000;">YV</span>NCARDDE<span style="color: #FF0000;">EQNL</span>VAFQYHR..Q<span style="color: #FF0000;">IFY</span>RTCRV<span style="color: #FF0000;">I</span>RPGC<span style="color: #FF0000;">ELLVWY</span>GDE<span style="color: #FF0000;">Y</span>GQELGIKWGSKWKKELMAGR | ||
3RAY PRDM11 <span style="color: #0066CC;">FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTS...GESDVRCVNEVIPKGHIFGPYEGQIS......TQDKSAGFFSWLIVDK.NNRYKSID.......GSDETKANWMRYVVISREEREQNLLAFQHSE..RIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLAR</span> | |||
2L9Z PRDM4 <span style="color: #0066CC;">WCTLCDRAYPSDCPEHGPVTFVPDTPIE....SRARLSLPKQLVLRQSIV..GAEVGVWTG.ETIPVRTCFGPLIGQQSHSMEVAEWTDKAVNHIWKIYHN.GVLEFCII.......TTDENECNWMMFVRKARNREEQNLVAYPHDG..KIFFCTSQDIPPENELLFYYSRDYAQQI</span>.............. | |||
3IXH PRDM10 WCEECNNAHASVCPKHGPLHPI<span style="color: #0066CC;">PNRPVL....TRARASLPLVLYIDRFLG......GVFSK.RRIPKRTQFGPVEGPLV.....RGSELKDCYIHLKVSLDKGDRKERDLHEDLWFELSDETLCNWMMFVRPAQNHLEQNLVAYQYGH..HVYYTTIKNVEPKQELKVWYAASYAEFVNQK</span>IHDISEEERK. | |||
3DAL PRDM1 <span style="color: #0066CC;">DGGTSVQAEASLPRNLLFKYATN.SEEVIGVMSK.EYIPKGTRFGPLIGEIY..TNDTVPKNANRKYFWRIYSR.GELHHFID.......GFNEEKSNWMRYVNPAHSPREQNLAACQNGM..NIYFYTIKPIPANQELLVWYCRDFAERL</span>HYPYPGELTMMNL. | |||
3JV0 PRDM2 LAEV<span style="color: #0066CC;">PEHVLRGLPEEVR.LFPSAVDKTRIGVWAT.KPILKGKKFGPFVGDKK.....KRSQVKNNVYMWEVYYP.NLGWMCID.......ATDPEKGNWLRYVNWACSGEEQNLFPLEINR..AIYYKTLKPIAPGEELLVWYNGEDNPEIA</span>AAIEEERASARSK | |||
3EP0 PRDM12 S<span style="color: #0066CC;">GEVQKLSSLVLPAEVIIAQSSIPGEGL.GIFSK.TWIKAGTEMGPFTGRVI..APEHVDICKNNNLMWEVFNEDGTVRYFID.......ASQEDHRSWMTYIKCARNEQEQNLEV.VQIGT.SIFYKAIEMIPPDQELLVWYGNSHNTFLG</span>IPGVPGLEEDQKK | |||
Phylogenetic variability of the knuckle-PR(SET) domain for PRDM7/9, shown below, is complicated by the various gene duplications of PRDM7. [http://genomewiki.ucsc.edu/index.php/Image:PRDMdifAlign.pdf Much less variability] occurs between the universally conserved patches in the other five genes with a comparable domain, namely PRDM11, PRDM4, PRDM10, PRDM15 and PRDM6. These genes did not experience duplications during placental evolution. The fact that the entire domain is strongly conserved -- with vary different amino acids in each protein -- implies strong selective pressure acts along the entire domain in these five proteins so the 3D structure is not floppy (indeterminate random coil) between the universally conserved patches, and that whatever functions these genes have remained constant during placental evolution. | |||
Note knuckle region in PRDM7/9 has moderate variability. Assuming on analogy with the terminal array zinc fingers that the residues between the second and third zinc ligands contain the residues that provide recognition specificity, these are <span style="color: #990099;">QNFFIDS</span>. This region has little phylogenetic variability in PRDM7/9. However overall PRDM11, PRDM4, PRDM10, PRDM15 and PRDM6 have [http://genomewiki.ucsc.edu/index.php/Image:knucklePhylo.pdf even less variability]. These regions could bind dna, single stranded rna or another protein involved in regulation. Those these partners may differ, the type of macromolecule will likely be the same because of underlying homology and implausibility of type change. The phylogenetic alignment of non-pseudogenes in the PRDM7/9 group is quite conservative from calJac (new world monkey Callithrix) to human: | |||
PRDM9_homSap Y<span style="color: #FF0000;">C</span>EM<span style="color: #FF0000;">C</span><span style="color: #0066CC;">QNFFIDS</span><span style="color: #FF0000;">C</span>AA<span style="color: #FF0000;">H</span>GPPTFVKDSAVDKGHPN<span style="color: #FF0000;">R</span>SAL<span style="color: #FF0000;">SLP</span>PGLRIGPSGIPQAGL<span style="color: #FF0000;">GV</span>WNEASDLPLGLH<span style="color: #FF0000;">FGP</span>YEGRITEDEEAANNGYS<span style="color: #FF0000;">WLI</span>TKGRNCYEY<span style="color: #FF0000;">VD</span>GKDKSWA<span style="color: #FF0000;">NWM</span>R<span style="color: #FF0000;">YV</span>NCARDDE<span style="color: #FF0000;">EQNL</span>VAFQYHRQ<span style="color: #FF0000;">IFY</span>RTCRV<span style="color: #FF0000;">I</span>RPGC<span style="color: #FF0000;">ELLVWY</span>GDE<span style="color: #FF0000;">Y</span>GQELGIKWGSKWKKELMAGR | |||
PRDM9_panTro .......................................................R.......P.....S.....Q.........S.............................E............S......S.......................................... | |||
PRDM9_gorGor ...................I.....................................................K........................................................................................................ | |||
PRDM9_ponAbe ................................................................................K......................................W.......................................................P.. | |||
PRDM9_nomLeu .....................I...T.G...........................................................................................W.......................................................P.. | |||
PRDM9_macMul .....................I.....E..................................................Q................................................................................................... | |||
PRDM9_papHam ...........................N....................................................K................................................................................................. | |||
PRDM7_homSap .....................................................................................S......................S..................................................................... | |||
PRDM7_ponAbe .....................................T........................................K................................................................................................... | |||
PRDM7_calJac ...I.............................HA.........................................V.......SS............................................................................................ | |||
PRDM7_micMur ...K.......................................K.R................E............QV........S....................D..............E...................Q.............................E..TIRQ | |||
PRDM7_otoGar ...K.......N..V.........T..E.......V....S..G.RT.......F.........Q..........QV........S....................E.QG...........E....................................................T..Q | |||
PRDM7_tupBel ...K.........S.....I..........SL...V.........A.....E.......A.T.............Q.........S....................E.C..............................................E................S.WQ.E | |||
PRDM9_oryCun ...K.....L....V....I...............V...............E.......................Q...E.....S....................R.............N............K.......Q..K..........................E..T... | |||
PRDM7_oryCun ...K.....L....V....I...............V...............E.......................Q...E.....S....................R.............N............K.......Q..K..........................E..T... | |||
PRDM7_ochPri ..........E...V..S..............H..V....S..........E........TT.............QV..E...T.S...........R........P.Q...........N.....................AV.Q.........................E..T... | |||
PRDM7_ratNor ...K.........PN....V.....V..R....H.V...............E.............V.......K.Q.........S..................Q.E.Q.........................K............R.....................M..GFT... | |||
PRDM7_musMus ...K.........PN....L.....M..R....H.V.........S.....E.............V.........Q.........S..................Q.E.Q.........................K..................................M..GFT... | |||
PRDM7_musMol ...K.........PN....L.....M..R....H.V.........S.....E.............V.........Q.........S..................Q.E.Q.........................K..................................M..GFT... | |||
PRDM7_dipOrd ...Q......N..TV....I..R.NV.....YD..V.........RQ.S..E........E..............Q.....D...S....M........V........Q...........Y.......................KA.........................R..T... | |||
PRDM7_speTri ..DK.....M...PV......I...V.N.D.S.H.T....L.......S..E.........T...........R.Q.........S....................E.Q.................................................................S... | |||
PRDM9_bosTau ..QE.........D.............E...A...V.T.....S.KL....E..........H............Q..D.K..I.S...........S........T.L...........HY...........G.......Q.V...............EK....CE.RG.SMFA... | |||
PRDM9_oviAri ..QE......N..D.............E...A.....T.....S.RL....E..........H............Q..D.K..V.S....................T.L....................L..QG.......Q.V................D....RD.SG.S..A... | |||
PRDM9_munMun ...E......N.............C..E...A.....T..H..S.RL....D.......KV...A........K.Q..DN.....S..A.................T..........................G.......Q.V................DF...RN.RG.S..A... | |||
PRDM7_turTru ...K.............A.........E.........T.....S.R.....E.......................Q.........S....................T..............E.....................V..............S.....P...G..SQ.V... | |||
PRDM7_lamPac ...K.................................T.......R.....E.........H.............QV........S..........K..........Y...............................................E................S.WQ.E | |||
PRDM7_susScr ...K.................................T.......R.....E.........H.............QV........S.........................................................V..............................T..I | |||
PRDM7_felCat ...K..........V.........N..G.........T.......R..S..E...............T.......Q.........S....................N................................................................S..ST.K | |||
PRDM7_ailMel ...K..........V...............Q......T.......R.............................Q.........S....................N..............E.................................................S..A..K | |||
PRDM7_pteVam ...K.............S.I.....E..IR.......T.............E................L......QV........S.....QG.............E.R..............................................................R..T... | |||
PRDM7_myoLuc ...K..........V................A.....T.............E........EC...V...Y.....Q.....AI..S....................T.Q..................................V.K.........E...............T.PV... | |||
PRDM7_equCab ...N...............I.................T..L....R.....E.......................Q.........S....................I....................................V...........................R..T... | |||
PRDM7_sorAra ...N......NK.S...S.I....N..A...S.....T..H..........E....I..................Q..N......S..................V.E.L............Y....................I.K..........................S..T.DK | |||
PRDM9_loxAfr ...K.......T..V..A.M.....P..R....H...T..........S..K..........E............QV...K....S.........K..........E..............E....................T.Q.D.....................R.....TS.. | |||
PRDM7_choHof ...K.....FEN........LL.....GQ.R.KH...V......L......E.......................QV......T.S......................C.................................A...............................T.EK | |||
PRDM9_homSap Y<span style="color: #FF0000;">C</span>EM<span style="color: #FF0000;">C</span><span style="color: #0066CC;">QNFFIDS</span><span style="color: #FF0000;">C</span>AA<span style="color: #FF0000;">H</span>GPPTFVKDSAVDKGHPN<span style="color: #FF0000;">R</span>SAL<span style="color: #FF0000;">SLP</span>PGLRIGPSGIPQAGL<span style="color: #FF0000;">GV</span>WNEASDLPLGLH<span style="color: #FF0000;">FGP</span>YEGRITEDEEAANNGYS<span style="color: #FF0000;">WLI</span>TKGRNCYEY<span style="color: #FF0000;">VD</span>GKDKSWA<span style="color: #FF0000;">NWM</span>R<span style="color: #FF0000;">YV</span>NCARDDE<span style="color: #FF0000;">EQNL</span>VAFQYHRQ<span style="color: #FF0000;">IFY</span>RTCRV<span style="color: #FF0000;">I</span>RPGC<span style="color: #FF0000;">ELLVWY</span>GDE<span style="color: #FF0000;">Y</span>GQELGIKWGSKWKKELMAGR | |||
=== Central PR(SET) domain descended from PRDM11 === | |||
Various additional sequences are relevant to understanding the curated placental mammal PRDM7/9 set. For example, the neanderthal genome despite being very far from satisfactory coverage can provide a PRDM9 sequence derived from the human reference sequence using non-synonymous SNPs reported in the corresponding UCSC browser track. The changes reported in the zinc finger domain (R HDL S R) may be enough to have created somewhat of a species barrier, though this involves comparing a fossil sequence to a contemporary human (which are today themselves quite variable). Similarly, the bushman genome sequence might yield an intermediate outgroup, though that assembly (like so many others) remains elusive. | |||
Terminal sequences for 9 additional species of murid rodents have been determined but these have limited value for comparative genomics because they do not even cover the entire terminal exon and their syntenic contexts (and thus homological relationships) were not established. The single individual sequenced may not be representative of the overall population in the zinc finger region (based on the extensive diversity observed in human), diminishing their utility for predicting species barriers. These genes are most likely PRDM7 orthologs only secondarily related to the catarrhine primate PRDM9 set, ie descended from the unique locus present in stem euarchontoglires whereas the latter duplicated from a stem old world monkey PRDM7. It is worth noting that the reported sequences are very orderly and lack the overall chaos of frameshifts and stop codons so often seen in this gene family. The protein accessions are [http://www.ncbi.nlm.nih.gov/protein/ADA68112,ADA68113,ADA68116,ADA68117,ADA68118,ADA68119,ADA68120,ADA68121,ADA68122 here]. | |||
A [http://www.uniprot.org/uniprot/Q6P2A1 zebrafish protein] put forward as an ortholog to placental mammal PRDM9 seems implausible given that birds, lizard and frog lack notable homologs. It lacks close counterparts in other species of fish with determined genomes and is not syntenic to mammalian gene locations. Thus it might represent an independent gene shuffle that resulted in a similar concatenation of domains (parallel evolution). | |||
The protein lacks the KRAB and SSXRD domains but contains a standard knuckle, PR(SET), early ZNF finger and ZNF repeat domain (all in exons phased identically to human). Although back-blast restricted exons 3-5 to the human proteome has best matches to PRDM9, PRDM7 and the closely related PRDM11 (suggesting orthology of this region), the pre-zinc finger part of exon 6 does not give a clear signal despite its early C2H2 domain, perhaps because of few conserved residues after it. The same could be said for the pre-zinc finger of PRDM9 -- it is apparently just a fast evolving linker region not under selection for amino acid sequence. | |||
While blastp of the zinc finger array is always a problematic exercise, here it gives closer resemblance to other zinc finger genes, notably ZNF658 and ZNF 585, than to any member of the PRDM family. The phase 12 intron here is moderately diagnostic in conjunction with the early zinc finger. The zebrafish terminal zinc finger array, while disorderly, does have several zinc fingers ending in the GEKP-like lockdown cap which supports a relationship with similar caps in PRDM7/9. Genes related to the zebrafish feature are found in salmon, trout, catfish and minnow but not stickleback, fugu, tetraodon or medaka. Transcripts are exceedingly common in contrast to mammals. The missing KRAB and SSXRD domains are believed critical in recruiting other essential proteins to the hotspot in the only systems with experimental data (mouse and human) so this gene cannot fulfill the same functional role. | |||
Thus only central PR(SET) exons are established as directly relevant to the history of PRDM7/9, ie that it was present in the common ancestor of mammals and fish. The terminal exon could represent orthology with extreme relative divergence but the evidence favors a chimeric origin with a different zinc finger terminal exon. The zebrafish gene is thus only a partially verifiable member of the PRDM family, one lacking a convincingly orthologous terminal exon as well as the final fusion with SSX1 as in PRDM7/9. Early diverging tetrapods need re-examination to see if they too have a gene with central PR(SET) exons of PRDM7/9 in a gene with a following phase 12 exon. | |||
Absence in frog, lizard and bird genomes would require persistence through the common ancestor but multiple independent loss events in the descendent lineages. Here frog still has a knuckle-PR(SET) domain most like that of PRDM7/9 but it is attached to a long BED domain and most resembles the human protein ZBED1 family overall. | |||
Even more attractive is the hypothesis that nearest neighbor PRDM11 -- which is highly conserved and effortlessly located in all tetrapods including frog, birds, lizard and platypus -- gave rise to PRDM7/9 via gene duplication. The duplicated gene subsequently neofunctionalized by reciprocal translocation with SSXRD and a ZNF gene to acquire its current N- and C-termini. PRDM11 has hardly changed since the parenting event. Best-blast (of ancestral, consensus, or any individual species) to human proteins is far and away PRDM7/9. This scenario explains -- without multiple gene loss events -- why PRDM7/9 cannot be located in early diverging tetrapods. | |||
[http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1000753 Reported] PRDM9 orthologs in early diverging bilatera such as Lottia, Capitella and Nematostella can be dismissed as independent occurrences of common ancient domain combinations. None of these domains are mammalian innovations -- PR(SET) traces back to bacterial methylases and zinc fingers also have a long and complex history. Without conservation of all mammalian domains, exon phasing, syntenic chromosomal location and demonstration of descent from a single gene in the last common ancestor, there is no basis for calling such genes orthologous nor assuming they function similarly in meiosis or illuminate mammalian PRDM7/9 evolution. Widespread expression in testes is not supportive as it conflicts with the very restrictive mammalian expression pattern. How could such a fundamental capacity be lost (and replaced by a non-homologous system) so many times in so many other lineages -- all of which have obligatory meiosis? | |||
>PRDM7_danRer Danio rerio (zebrafish) Q6P2A1 transcript BC064665 no KRAB SSXRD or exon 5 but <span style="color: #0066CC;">knuckle</span> <span style="color: #990099;">SET</span> <span style="color: #6699FF;">early ZNf</span> <span style="color: #FF0000;">C2H2 array</span> | |||
0 MSLSP 1 | |||
2 DLPPSEEQNLEIQGSATNCYSVVIIEEQDDTFNDQPF 1 | |||
2 <span style="color: #0066CC;">YCEMCQQHFIDQCETHGPPSFTCDSPA</span>ALGTPQRALLTLP<span style="color: #990099;">QGLVIGRSSISHAGLGVFNQGQTVPLGMHFGPFDGEEISEEKALDSANSWV 0 | |||
0 ICRGNNQYSYIDAEKDTHSNWMK 2 | |||
1 FVVCSRSETEQNLVAFQQNGRILFRCCRPISPGQEFRVWYAEEYAQG</span>LGAIWDKIWDNKCISQ 1 | |||
2 GSTEEQATQN<span style="color: #6699FF;">CPCPFCHYSFPTLVYLHAHVKRTH</span>PNEYAQFTQTHPLESEAHTPITEVEQCLVASDEALSTQTQPVTESPQEQISTQNGQPIHQTENSDEPDASDIYTAAGEISDEI | |||
HA<span style="color: #FF0000;">C</span>VD<span style="color: #FF0000;">C</span>GRSFLRSCHLKR<span style="color: #FF0000;">H</span>QRTI<span style="color: #FF0000;">H</span>SKEKP | |||
YC<span style="color: #FF0000;">C</span>SQ<span style="color: #FF0000;">C</span>KKCFSQATGLKR<span style="color: #FF0000;">H</span>QHT<span style="color: #FF0000;">H</span>QEQEKNIESPDRPSDI | |||
YP<span style="color: #FF0000;">C</span>TK<span style="color: #FF0000;">C</span>TLSFVAKINLHQ<span style="color: #FF0000;">H</span>LKRH<span style="color: #FF0000;">H</span>HGEYLRLVESGSLTAETEEDHT | |||
EV<span style="color: #FF0000;">C</span>FDKQDPNYEPPSRGRKSTKNSLKGRGCPKKVAVGRPRGRPPKNKNLEVEVQKIS | |||
PI<span style="color: #FF0000;">C</span>TN<span style="color: #FF0000;">C</span>EQSFSDLETLKT<span style="color: #FF0000;">H</span>QCPRRDDEGDNVEHPQEASQ | |||
YI<span style="color: #FF0000;">C</span>GE<span style="color: #FF0000;">C</span>IRAFSNLDLLKA<span style="color: #FF0000;">H</span>ECIQQGEGS | |||
YC<span style="color: #FF0000;">C</span>PH<span style="color: #FF0000;">C</span>DLYFNRMCNLRR<span style="color: #FF0000;">H</span>ERTI<span style="color: #FF0000;">H</span>SKEKP | |||
YC<span style="color: #FF0000;">C</span>TV<span style="color: #FF0000;">C</span>LKSFTQSSGLKR<span style="color: #FF0000;">H</span>QQS<span style="color: #FF0000;">H</span>LRRKSHRQSSALFTAAI | |||
FP<span style="color: #FF0000;">C</span>AY<span style="color: #FF0000;">C</span>PFSFTDERYLYK<span style="color: #FF0000;">H</span>IRR<span style="color: #FF0000;">H</span>HPEMSLKYLSFQEGGVLSVEKP | |||
HS<span style="color: #FF0000;">C</span>SQ<span style="color: #FF0000;">C</span>CKSFSTIKGFKN<span style="color: #FF0000;">H</span>SCFKQGEKV | |||
YL<span style="color: #FF0000;">C</span>PD<span style="color: #FF0000;">C</span>GKAFSWFNSLKQ<span style="color: #FF0000;">H</span>QRI<span style="color: #FF0000;">H</span>TGEKP | |||
YT<span style="color: #FF0000;">C</span>SQ<span style="color: #FF0000;">C</span>GKSFVHSGQLNV<span style="color: #FF0000;">H</span>LRT<span style="color: #FF0000;">H</span>TGEKP | |||
FL<span style="color: #FF0000;">C</span>SQ<span style="color: #FF0000;">C</span>GESFRQSGDLRR<span style="color: #FF0000;">H</span>EQK<span style="color: #FF0000;">H</span>SGVRP | |||
CQ<span style="color: #FF0000;">C</span>PD<span style="color: #FF0000;">C</span>GKSFSRPQSLKA<span style="color: #FF0000;">H</span>QQL<span style="color: #FF0000;">H</span>VGTKL | |||
FP<span style="color: #FF0000;">C</span>TQ<span style="color: #FF0000;">C</span>GKSFTRRYHLTR<span style="color: #FF0000;">H</span>HQKM<span style="color: #FF0000;">H</span>S* 0 | |||
>ZBED1_xenTro | |||
0 MQAAEEACAQLEDELL 1 | |||
2 <span style="color: #0066CC;">FCEDCRLYFRDSCPTHGAPTFILDTPV</span>PENVPSRALLSLPEGLVVKERP<span style="color: #990099;">QGGFGVWCTIPVIPRGCIFGPYEGDVIMDRSDCTVYSWA 0 | |||
0 VRENGSYFYIDASDDSKSSWMR 2 | |||
1 YVACASTEEEHNLTVFQYRGKIYYRASQVIPTGTELLVWIGEEYARTLG</span>LKL 1 | |||
2 GEHFKYEFGEKELLMKLFQDLQLKPVDSISNHVSSQSQYMCNDMVTPVMQAHRTSYPLNNIGHTSSVFPLLEGTQNLVSLGRAQSRYWTFFGFQGDAYGRIIDKTK<span style="color: #6699FF;">IICKLCGVRLSYSGNTTNLRQHLIYK</span>HRRQYNDL | |||
>PRDM11_conSeq consensus of 30 tetrapod PRDM11 orthologs | |||
2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKEASGENDVRCINEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 | |||
0 IVDKNNRYKSIDGSDETKANWMR 2 | |||
1 YVVISREEREQNLLAFQHSERIYFRACRDIRPGERLRVWYSEDYMKRLHSMSQETIHRNLAR 1 | |||
PRDM9_homSap YL Y..M..NF.I.S.AA....T..K.SA.DK.H.N.S..SL.P.LRIGPSGIPQAGLGVW..AL.L.LH......R.TEDEEA.NNY... .TKGR.C.EYV..K.KSW..... ..NCA.DDE....V...YHRQ.FY.T..V....CE.L...GDE.GQE.GIKWGSKWKKE.MA | |||
PRDM11_homSap FW FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTSGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWL IVDKNNRYKSIDGSDETKANWMR YVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLAR | |||
PRDM11_panTro .. ........................................................................................ ....................... .............................................................. | |||
PRDM11_rheMac .. .........................................................I.............................. ....................... .............................................................. | |||
PRDM11_calJac .. ........................................................................................ ....................... ....................C......................................... | |||
PRDM11_otoGar .. ............................M..................EA...N....I.............................. ....................... ..A...............................R........................... | |||
PRDM11_musMus .. ................................................AG.......I.............................. ....................... ..................................R........................... | |||
PRDM11_ratNor .. ..............T................................EVG.......I...V.......................... ....................... ..................................R........................... | |||
PRDM11_cavPor .. .............................................I.EAG.......I.............................. ...............D......- ..................................R........................... | |||
PRDM11_speTri .. ...............................................EA........IS............................. ....................... ..........R.......G...............R......Q...R................ | |||
PRDM11_oryCun .. ..........................................L...QEA........I.D.....R.........AA........... .....S................. ........Q.........N.H..........A..R......G.................... | |||
PRDM11_ochPri .. ............................M..A...............EA........LSD............................ ....................... ....................H.........Q...R........................... | |||
PRDM11_bosTau .. ...............................................EA...N....I.......R...................... ....................... ........S.........................R........................... | |||
PRDM11_equCab .. ...............................................EA...N....I.......................T...... ....................... ..................................R........................... | |||
PRDM11_canFam .. ...............................................EA...N....I.............................. ....................... ..................................R......................H.... | |||
PRDM11_myoLuc .. ..............K....M..........L.................AN..N....I.............................. ....................... ....................H.......................................T. | |||
PRDM11_pteVam .. ................................................A...N...SI.............................. ................S...... ....................H.............R........................... | |||
PRDM11_eriEur .. ..............K...........................V....EA........I.............................. ......H.........S...... ....................H.............R........................... | |||
PRDM11_loxAfr .. ...............................................EA...N....IS............................. ..........V............ .........................V........R......Q.................... | |||
PRDM11_echTel .. ............................M..................EG...N....IS....T-..LR......Y.RN......... ....................... ....C...............H.............R.....GQ..................T. | |||
PRDM11_dasNov .. ...............................................EA...N....I.............................. ....................... .........................S........Q...............V........... | |||
PRDM11_macEug .. ............................M...........P......EA..QN....M.............................. .............T...Q..... ..I..........M......K....V........R.....................Q...T. | |||
PRDM11_monDom .. ............................M...........P......EA..Q.....M.............................. ......H......T......... .............M......K....V........R.....................Q...T. | |||
PRDM11_ornAna .. ........................................P.I....EA...N....M.............................. .............T......... .I.......................V........R.........................T. | |||
PRDM11_galGal .. ........................................P.I....EP...N....M..................S........... .............T......... ..I..........M....................K.....................N...TT | |||
PRDM11_taeGut .. ........................................P......EP...N....M..................S..R........ .............T......... ..I..........M................H...K....................MN.SFTS | |||
PRDM11_anoCar .. ...................M.L..A...I.........V.P......EAN..R.....G.I....R.Y.....KL.S........... .............T...TS.... ..A..........M...........T........R.....................N...T. | |||
PRDM11_xenTro .. ..............S....IL.P..L..I.M.E....SV.C.I.....S...R.....G.I....R.Y.....KL.S........... .............T...TS.... .............M......K....T....Q...K.....................N...TQ | |||
=== Structural considerations in C2H2 zinc fingers === | |||
High resolution structures of C2H2 zinc finger domains have been available for decades. As the name suggests, the divalent zinc atom locks the two cysteines and two histidines into a rigid geometry providing a core conformation that a small peptide of 28 residues could not otherwise stably assume. Note in the unbound state, finger tips must retain flexibility while the domain ensemble scans its genome for specific dna sequences appropriate to its function. Each finger binds a trinucleotide -- in effect making a zinc finger the protein counterpart to tRNA anticodon. However overall binding is not a simple read-off code because adjacent fingers alter each other's specificities in subtle ways. | |||
The linker region TGEKP plays a key role when the correct DNA sequence is encountered, [http://www.ncbi.nlm.nih.gov/pubmed/10656784 snap-locking] its finger down onto its target by capping the C-terminus of its alpha helix. A hydrogen bond between the first threonine and middle glutamate is key to this binding-induced conformational shift. From comparative genomics, it appears that a serine in first position can also form this hydrogen bond. The role of the glycine is to stay out of the way; the lysine counterbalances the negative charge of the glutamate; the proline terminates any helical propensity, allowing a fresh start in the adjacent finger. | |||
While this motif is immensely conserved within C2H2 zinc finger of PDRM9 homologs, exceptions do occur. It is important to understand these because these loss of dna lock-down could loosen or even eliminate trinucleotide binding specificity. Such steps might represent initial stages of pseudogenization. However many exceptions occur within the first or last fingers. It is also common for fragmentary and imperfect motifs to end the protein, sometimes continuing on in another reading frame past the current stop codon. | |||
Note in aligning zinc finger motifs, the breaks should always be put at the end of the linker region. It is completely illogical to break at the first cysteine as some authors do because capping by the linker region is specific to its zinc finger, not the following one. | |||
=== Predicting dna binding sites of zinc finger domains === | |||
[[Image:PRDM9onDNA.jpg|left]] | |||
<br clear = all> | |||
== Origin of Species and all that == | |||
The origin of species is an old and exceedingly complex topic. PRDM7/9 cannot provide a unified molecular basis for it because orthologs do not exist outside of mammals. Yet genetic mapping shows PRDM7 at the core of hybrid sterility in mice (a enticing proxy for speciation) and PRDM9 sites are key to meiotic recombination in humans. | |||
Although placental mammals very greatly in the specifics of PRDM7/9 gene expansion, some variation on the mouse/human might still be applicable. However that cannot be the sole explanation even here because some species such as dog lack any functional gene family member. An [http://genome.cshlp.org/cgi/pmidlookup?view=long&pmid=22006216 Oct 2011 paper] established that the PRDM7 gene of the boxer used for the genome project is in fact representative of dog breeds; wolves, jackals and foxes also have the same inactivating frameshifts. Other carnivora such as mink -- while possessing a seeminly intact PRDM7 gene -- lack a sufficiently long zinc finger array to specify hotspots. Some other mechanism is needed to mark the sites of double stranded breaks that initiate meiosis in these Carnivora. | |||
It is problematic whether the PRDM7/9 mechanism extends to marsupials and monotremes and not an option for earlier amniotes such as birds which lack any semblance of the gene (though homologs for its parts occur). Conceivably, a zinc finger array from a different gene family steps in. However drosophila utilizes very different gene products to control meiosis (with other genes such as DMRT1 more universal). There cannot be a unified molecular basis for speciation within bilaterans based on zinc finger proteins. | |||
Thus despite being an ancient core process, the meiotic machinery is unexpectedly not conserved at the gene family level, instead exhibiting discontinuities in some gene families utilized to accomplish it. That is not unlike SRY sex determination (which underwent abrupt changes in marsupials and placentals shortly after divergence from monotremes), or the sex chromosomes themselves (which are homologous to bird but with hetergametic XY males in platypus and taken from different autosomes in therans). | |||
PRDM7/9 do affect an immense number of other important topics in mammalian evolution through their specification of meiotic recombination sites. Because the crossover bottleneck in pseudoautosomal regions, these influences become deeply intertwined with special aspects of sex chromosome evolution. Moving beyond the narrow perspective of PRDM7/9 evolution, the following issues can be explored: | |||
* meiotic sex chromosome inactivation (MSCI) | |||
* progressive chromosome Y degeneration or conversely maintainence | |||
* gene dosage compensation for chromosome X | |||
* sex determination via SOX3 evolving into SRY on chromosome Y after platypus divergence | |||
* co-evolution with key meiotic regulatory genes such as DMRT1 | |||
* expansion and contraction of the pseudoautosomal region in placentals | |||
* barriers to introgression during hominid history | |||
* impacts on lineage sorting within great apes | |||
* boundaries and persistence of haploblocks | |||
* localization of gene conversion | |||
* random X-inactivation in females | |||
* XY bodies and chromatin structure during meiosis | |||
* evolutionary strata in sex chromosomes due to inversions reducing recombining region | |||
* translocations and other chromosomal rearrangements in evolution and disease | |||
> | These topics will be further developed in October. | ||
0 | |||
0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | (to be continued) | ||
2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | |||
2 | == Supplemental information == | ||
0 | |||
2 | The sections below store data used above. This includes curated sequences from all available mammals for PRDM7 and PRDM9 and additional their partial paralogs in the PRDM gene family. These latter have extensive comparative genomics alignments readily available elsewhere (UCSC genome browser, under GeneSorter feature and ProteinFasta feature in gene details page) so that is not repeated here. | ||
2 | |||
While this topic has a long history in the peer-reviewed scientific literature, only the most recent articles are provided here because their reference sections satisfactorily summarize pre-2005 studies. Instead, the focus here is identifying free full text access to the recent articles, preferably as html which better supports copying snippets of text. The journal, google, and PubMed all provide forward citations to still other articles that cite the articles provided here. | |||
=== Curated reference sequences === | |||
The sequences below have largely been compiled from genome projects -- only rarely do validating transcripts exist at GenBank. Sequences with a single frameshift or other glitch have been edited in some cases to show full length proteins on the theory that the error either reflects an atypical individual chosen for sequencing, sequencing error in low coverage projects within a difficult region, one allele in balanced polymorphism, or a mutant allele. However such sequences may instead reflect early stages of pseudogenization. Other sequences are in fact clearly pseudogenes; here recognizable exons are represented to allow determination of historic repeat number and rough dating of loss of function. | |||
Chimp PRDM9 has a thoroughly garbled 11th repeat (3 frameshifts, nnnnn in the CGSC 2.1.3/panTro3 Oct 2010 assembly) followed by eight additional repeats. It is difficult to say if these are still in the initial reading frame. No data is currently available for Pan paniscus but despite minimal coverage, Pan troglodytes schweinfurthii has a long trace read in this region (ti|2009092447) that covers repeats 3-11 sharing the first frameshift in Pan troglodytes but soon degenerating into unmistakable pseudogene (ie the frameshifts in Pan troglodytes are not sequencing artifacts): | |||
PRDM9 Pan troglodytes schweinfurthii | |||
THTGEKP 3 | |||
YVCRECGRGFSVKSSLLSHQSTHTGEKP 4 | |||
YVCRECGRGFSVKSSLLSHQRTHTGEKP 5 | |||
YVCRECGRGFSVKSSLLSHQRTHTGEKP 6 | |||
YVCRECGRGFSVKSSLLSHQRTHTGEKP 7 | |||
YVCRECGRGFSQQSHLLSHQRTHTGEKP 8 | |||
YVCRECGRGFSQQSHLLSHQRTHTGEKP 9 | |||
YVCRECGRGFSQQSHLLSHQRTHTGEKL 10 | |||
YVCRECGRGFSRAVTPPQTPETHTGEKL 11 | |||
YVCRE<font color=magenta>c</font>GRGFSDK<font color=magenta>s</font>SLlSVTRVHTQGRA 12 | |||
YVCRECGRGFSWQS 13 | |||
Gorilla, despite its importance, has a poor quality second assembly that is riddled with gaps and misplaced contigs. The PRDM9 gene terminates early in the zinc finger array with a small gap before a misplaced LINE element. The neighboring genes are not plausible though PRDM9 is correctly placed on chr5. PRDM7 in gorilla has four frameshifts in the last exon. Those are 'restored' below to reveal gene history (which in this case is 3.5 repeats with no trailing debris). Lower case letters indicate ends of correct reading frames. | |||
Sumatran orangutan PRDM9 has a distal zinc finger array frameshift in contig ABGA01214983 (<font color = red>a</font>GGGGAGAAG CCCTATGTCT GCAGGGAGTG) which also appears in the July 2007 assembly provided by UCSC (WUGSC 2.0.2/ponAbe2). However there is no support whatsoever for this extra adenosine in the underlying raw Sanger trace reads used to make the contig -- all reads omit it and have the expected reading frame. Consequently, the assembly appears to be in error. The full length sequence is provided here. Orang PRDM9 has the expected syntenic location between CDH10 and CDH12. | |||
Gibbon has a stop codon in exon 6 that is supported by all 6 of the available Sanger traces. | |||
In the case of more intensively studied species such as human and mouse, the number of C2H2 repeats varies widely. Only the reference sequence representative is shown here. This variation likely occurs in all species with the individual animal chosen for sequencing not necessarily the most common allele. Many clades have independent histories of gene amplification and gene loss, making both orthologous and functional comparisons problematic at substantial divergence. | |||
These sequences are under constant revision as anomalies surface and are resolved by re-examining the original Sanger trace reads. However when coverage is low, it is very difficult to distinguish read error from actual oddities. For example, the horse genome assembly shows a disabling frameshift in seventh exon. That is apparently based solely on trace ti|1206069852 and arises from a truncated homopolymer run error CCCC that is frame-preserving CCCCC in ti|1330418597, ti|1322386025 and ti|1288100157. These latter reads however later have problems of their own. Another issue is individual traces covering part of a zinc finger repeat -- these cannot always be assigned reliably to PRDM7/9 as many similar zinc finger repeats exist in these genomes. | |||
Other useful sequences such as PRDM11, PRDM4 and zinc finger semi-homologs having similar exon and domain structures, are provide in the subsequent section along with syntenic markers such as GAS8. | |||
>PRDM9_homSap Homo sapiens (human) genome Prim gene 13 CDH12 chr5 10 exon size 18,301 bp KRAB SSXRD SET C2H2 | |||
0 MSPEKSQEESPEEDTERTERKPM 0 | |||
0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | |||
2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | |||
2 VKPPWMALRVEQRKHQK 0 | |||
0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 | |||
2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1 | |||
2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 | |||
0 ITKGRNCYEYVDGKDKSWANWMR 2 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | ||
2 | 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK | ||
VKYGECGQGFSVKSDVITHQRTHTGEKL | |||
YVCRECGRGFSWKSHLLIHQRIHTGEKP | |||
YVCRECGRGFSWQSVLLTHQRTHTGEKP | |||
YVCRECGRGFSRQSVLLTHQRRHTGEKP | |||
YVCRECGRGFSRQSVLLTHQRRHTGEKP | |||
YVCRECGRGFSWQSVLLTHQRTHTGEKP | |||
YVCRECGRGFSWQSVLLTHQRTHTGEKP | |||
YVCRECGRGFSNKSHLLRHQRTHTGEKP | |||
YVCRECGRGFRDKSHLLRHQRTHTGEKP | |||
YVCRECGRGFRDKSNLLSHQRTHTGEKP | |||
YVCRECGRGFSNKSHLLRHQRTHTGEKP | |||
YVCRECGRGFRNKSHLLRHQRTHTGEKP | |||
YVCRECGRGFSDRSSLCYHQRTHTGEKP | |||
YVCREDE | |||
> | >PRDM9_panTro Pan troglodytes (chimp) 2010 assembly exon 8 in gap, 11th repeat 3 frameshifts, only 1 trace supports stop codon *VCREDE ti|451784509 | ||
0 | 0 MSPERSQEESPEEDTERTERKPM 0 | ||
0 | 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | ||
2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPLMALRVEQRKHQK 0 | ||
0 | 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 | ||
2 | 2 ELKKKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 | ||
2 | 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYKGRITEDEEAANNGYSWL 0 | ||
0 ITKGRNCYEYVDGKDKSWANWMR 2 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | ||
2 | 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPRSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTAKLFVGVGISRIAK | ||
VKYGECGQGFSVKSDVITHQRTHTGEKP | |||
YVCRECGRGFSWKSHLLSHQRTHTGEKP | |||
YVCRECGRGFSVKSSLLSHRTTHTGEKP | |||
YVCRECGRGFSVKSSLLSHQRTHTGEKP | |||
YVCRECGRGFSQQSNLLSHQRTHTGEKP | |||
YVCRECGRGFSVKSSLLSHQRTHTGEKP | |||
YVCRECGRGFSVKSSLLSHQRTHTGEKP | |||
YVCRECGRGFSKQSHLLSHQRTHTGEKP | |||
YVCRECGRGFSVQSNLLSHQRTHTGEKL | |||
YVCRECGRGFSQQSHLLRHQRTHTGEKP | |||
YVCR<font color=magenta>e</font>CGR<font color=magenta>g</font>FSV<font color=magenta>k</font>SSLLSHQRTHTGEKP | |||
YVCRECGRGFSVKSSLLSHQRTHTGEKP | |||
YVCRECGRGFSKQSHLLSHQRTHTGEKP | |||
YVCRECGRGFSQQSHLLSHQRTHTGEKP | |||
YVCRECGRGFSQQSHLLRHQRTHTGEKP | |||
YVCRECGRGFSVKSSLLSHQRTHTGEKP | |||
YVCRECGRGFSVKSSLLSHQRTHTGEKP | |||
YVCRECERGFSQQSHLLRHQRTHTGEKP | |||
YVCRECGRGFSRQSALLIHQRTHTGEKP | |||
<font color = red>*</font>VCREDE | |||
> | >PRDM9_gorGor Gorilla gorilla (gorilla) CABD02290262 CABD02290264 cdh12 chr5 ZNF at end of contig Aug 2009 assembly poor quality | ||
0 | 0 MSPERSQEESPEEDTERTERKPM 0 | ||
0 | 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | ||
2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPCMALRVEQRKHQK 0 | ||
0 | 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 | ||
2 | 2 ELRKKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 | ||
2 | 2 YCEMCQNFFIDSCAAHGPPIFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYKGRITEDEEAANNGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | ||
2 | 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARTLLQPENPCPGDQNQEQQYPDPRSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK | ||
VKYGECGQGFSVKSDVITHQRTHTGEKP | |||
YVC | |||
>PRDM9_ponAbe Pongo abelii (Sumatran orangutan) CDH10- PRDM9+ CDH12- distal frameshift aGGGG --> GGGG causing loss of 1.5 repeats in ABGA01214983 | |||
0 MSPERSQEESPEDDTERTERKPT 0 | |||
> | |||
0 | |||
0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | ||
2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPWMALRVEQRKHQK 0 | ||
0 | 0 GMPKASFNNESSLKELSETANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 | ||
2 | 2 ELRSKETEGNTYSLRERKGHAYKEISEPQDDDYL 1 | ||
2 | 2 CEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALTLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITKDEEAANNGYSWL 0 | ||
0 ITKGRNCYEYVDGKDKSWANWMR 2 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | ||
2 | 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNHEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK | ||
VKYGECGQGFSVKSDVITHQRTHTGEKP | |||
YVCRECGRGFSRQSVLLIHQRTHTGEKP | |||
YVCRECGRGFSRRSVLLIHQRTHTGEKP | |||
YVCRECGRGFSQQSVLLIHQRTHTGEKP | |||
YVCRECGRGFSRRSVLLIHQRTHTGEKP | |||
YVCRECGRGFSWKSVLLRHQRTHTGEKP | |||
YVCRECGRGFSQQSVVFIHQRTHTGEKP | |||
YVCRECGRGFSGKSVLFRHQRTHTGEKP | |||
YVCRECGRGFSDKSGVCYHQRTHT<font color = magenta>g</font>EKP | |||
YVCRECGRGFSVKSNLLSHQRTHTEEKL | |||
YVCREDE* | |||
> | >PRDM9_nomLeu Nomascus leucogenys (gibbon) ADFV01015315 Prim gene 10 cdh12 ADFV01015317 ADFV01015319 no synteny CpG stop exon 6 in 6/6 traces VCRKDE* in altered reading frame | ||
0 | 0 MSPERSQEESPEEDTERTEQKPT 0 | ||
0 | 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | ||
2 | 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPWMALRMEQRKHQK 0 | ||
0 | 0 GMPKASFSNESSLKELSGAANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 | ||
2 | 2 ELRRKETEGKMYSL<font color = red>*</font>ERKGHAYKEVSEPQDDDYL 1 | ||
2 | 2 YCEMCQNFFTDSCAAHGPPTFIKDSTVGKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCQVIRPGCEPLVWYGDEYGQELGIKWGSKWKKELTAER 1 | ||
2 | 2 EPKAEIHPCPSCCLAFSSQKFLSQHVARHHSSQNFPGPSARKFLQPENPCPGDQNQEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWQREISRAFSSSPKVQMGSCRVGKRIIEEESRTGQKVNPGNTGQLFVGVGISRIAE | ||
VKYAECGQGFSDKSDVITHQRTDTGEKP | |||
YLCRECGRGFSVKSSLLSHQRTHTGEKP | |||
YVCRECGRGFSKKSNLLSHQRTHTGEKP | |||
YVCRECGRGFSDKSSLLRHQRTHTGEKP | |||
YVCRECGRGFSQKSSLLSHQRTHTGEKP | |||
YVCRECGRGFSQKSSLLSHQRTHTGEKP | |||
YVCRECGRGFSDKSSLLRHQRTHTGEKP | |||
YVCRECGRGFSQKSSLLSHQRTHTGEKP | |||
YVCRECGRGFSVKSNLLSHQRTHTGEKP | |||
YVCRECGRGFSDKSSLLRHQRTHTGEKP | |||
<font color = red>*</font><font color = magenta>v</font>CRKDE | |||
> | >PRDM9_macMul Macaca mulatta (rhesus) genome Prim gene 9 CDH12 chr6 exon 4 lost to Ns | ||
0 | 0 MSPERSQEESPEEDTERTERKPT 0 | ||
0 | 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | ||
2 | 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPWMAVRVEQSKHQK 0 | ||
0 | 0 GMPKASFNNESSLKEVSGMANLLNTSGSEQAQKPVSPPGEARTSGQHSRLKL 1 | ||
2 | 2 ELRRKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 | ||
2 | 2 YCEMCQNFFIDSCAAHGPPTFIKDSAVEKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITQDEEAANNGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | ||
2 | 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSTQNFPGPSARRLFQPENLCSGDQNQEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWPKEISRAFSSPPKGQMGSSRVGERMMEEEYRTGQKVNPENTGKLFVGVGISRIAK | ||
VKYGECGQGFSDKSDVIIHQRTHTGEKP | |||
YLCRECGRGFSQKSSLRRHQRTHTGEKP | |||
YLCRECGRGFRDNSSLRYHQRTHTGEKP | |||
YLCRECGRGFSNNSGLCYHQRTHTGEKP | |||
YLCRECGRGFSDNSSLHRHQRTHTGEKP | |||
YLCRECGRGFSNNSGLRYHQRTHTGEKP | |||
YLCRECGRGFSNNSGLRHHQRTHTGEKP | |||
YLCRECGRGFSQKANLLRHQRTHTGEKP | |||
YLCRECGRGFSQKADLLSHQRTHTGEKP | |||
<font color = red>*</font><font color = magenta>v</font>CRKDE | |||
>PRDM9_macFas Macaca fascicularis (crab-eating macaque) CAEC01530962 CAEC01530970 frameshift exon 7 fragmentary array | |||
0 MSPERSQEESPEEDTERTERKPT 0 | |||
0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | |||
2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | |||
0 GMPKASFNNESSLKEVSGMANLLNTSGSEQAQKPVSPPGEARTSGQHSRLKL 1 | |||
2 ELRRKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 | |||
2 YCEMCQNFFIDSCAAHGPPTFIKDSAVEKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHF<font color = magenta>g</font>PYEGRITQDEEAANNGYSWL 0 | |||
0 ITKGRNCYEYVDGKDKSWANWMR 2 | |||
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | |||
2 EPKPEIHPCPSCCLAFSSQKFLSHHVERNHSTQNFRGPSARRLLQPENLcSGDQNQEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWQKEISRAFSSPPKGQMGSSRVGERMMKEEYRTGQKVNPENTGKLFVGVGISRIAQ | |||
VKYGECGQGFSEKSDVIIHQRTHTGEKP | |||
YLCRECGRGFSRKSNL | |||
> | >PRDM9_papHam Papio hamadryas (baboon) genome Prim gene 11 cdh12 contigs scattered | ||
0 0 | 0 0 | ||
0 1 | 0 1 | ||
2 | 2 1 | ||
2 | 2 VKPPWMAFRVEQSKHQK 0 | ||
0 | 0 EMPKTSFSNESSLKELSGTPNLLSTSGSEQAQKPASPPGEASTSGQHSRLKL 1 | ||
2 | 2 ELRRKEAEGKMYSLRERKGHAYKEVSELQDDDYL 1 | ||
2 | 2 ycEMCQNFFIDSCAAHGPPTFVKDSAVNKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDKEAANNGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 1 | ||
2 | 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSTQNFPGPSARRLLQPENLCSGDQNQEQQYSDPCSCNDKTKGQEIKERSKLLNKRTWQKEISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPENIGKLFVEVGISRIAK | ||
VKYGECGQGFSDKSDVVIHQRTHTREKP | |||
YLCRECGRGFSQKSNLLRHQRTHTGEKP | |||
YLCRECGRGFRDNSSLRCHQRTHTGEKP | |||
YLCRECGRGFRDNSSLRCHQRTHTGEKP | |||
YLCRECGRGFSDNSSLRYHQRTHTGEKP | |||
YLCRECGRGFRDNSSLRYHQRTHTGEKP | |||
YLCRECGRGFSVKSNLLSHQRTHTGEKP | |||
YVCRECGRGFSDNSSLRCHQRTHTGEKP | |||
YLCRECGRGFSQMSHLRCHQRTHTGEKP | |||
YLCRECGRGFSVKSNLLSHQRTHTGEKP | |||
YVCRECGRGFSRKANLLSHQRTHTGEKP | |||
<font color = red>*</font><font color = magenta>v</font>CRKDE | |||
> | >PRDM7_homSap Homo sapiens (human) pseu chr16 TUBB3+ DEFB+ AFG3L1+ DBNDD1- GAS8+ PRDM7- 4 frameshifts | ||
0 | 0 MSPERSQEESPEGDTERTERKPM 0 | ||
0 | 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKMNYNALITV 1 | ||
2 | 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPWMAFRGEQSKHQK 0 | ||
0 | 0 GMPKASFNNESSLRELSGTPNLLNTSDSEQAQKPVSPPGEASTSGQHSRLKL 1 | ||
2 | 2 ELRRKETEGKMYSLRERKGHAYKEISEPQDDDYL 1 | ||
2 | 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSSANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWSGDEYGQELGI<font color = magenta>k</font>WGSKWKKELMAGR 1 | ||
2 | 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQERQYSDPRCCNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK | ||
VKYGECGQGFSDKSDVITHQRTHTGGKP | |||
YVCRECGR<font color = magenta>g</font>FSRKSDLLSHQRTHTGEKP | |||
YVCRECERGFSRKSVLLIHQRT<font color = magenta>h</font>TGETP | |||
<font color = magenta>v</font>CRKDE | |||
> | >PRDM7_panTro Pan troglodytes (chimp) genome Prim pseu 2 GAS8+ chr16 | ||
0 | 0 MSPERSQEESPEEDTERTERKPM 0 | ||
0 | 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | ||
2 | 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPLMALRVEQRKHQK 0 | ||
0 | 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 | ||
2 | 2 ELKKKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 | ||
2 | 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYKGRITEDEEAANNGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | ||
2 | 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLQPENP<font color = red>*</font>PGDQNQERQYSDPRCCNDKTKGQEVKERSKLLNKWTWQREISRAFSSLPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK | ||
VKYGECGQGFSDKSDVITHQRTHTGGKP | |||
YVCRECGQGFSRKSVLLIHQRTHRGEKP | |||
<font color = red>*</font><font color = magenta>v</font>CRKDE | |||
> | >PRDM7_gorGor Gorilla gorilla (gorilla) genome Prim pseu 3 GAS8+ chr15730 four frameshifts in last exon | ||
0 | 0 MSPERSQEESPEGDTERTERKPM 0 | ||
0 | 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | ||
2 | 2 GLRATQPVFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 0 | ||
0 | 0 GMPKASFNNESSLKELSGTPNLLNTSGSEQAQKPVSPPGEASTSGQHSRRKL 1 | ||
2 | 2 ELRRKETEGKMYSLRERKGHAYKEISKPQDDDYL 1 | ||
2 | 2 yCEMCQNFFIDSCAAHGPPTFVKDSAVDKRHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDKEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVALQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGR 1 | ||
2 | 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQERQYSDPRCCNDKTKGQEIKERSKLLNK<font color = magenta>r</font>TWQR<font color = magenta>e</font>ISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK | ||
YGECGQGFSWKSNLLRHQRTHTGGKP | |||
YVCRECGRGFSWKSDLLSHQRTHTGEKP | |||
YVCRECGRGFSWKSNLLSHQRTHT<font color = magenta>g</font>EKP | |||
<font color = red>*</font><font color = magenta>v</font>CRKDE | |||
> | >PRDM7_ponAbe Pongo abelii (Sumatran orangutan) genome-blat same late stop codon as chimp *VCRKDE* no cryptic repeats, adjacent GAS8 | ||
0 | 0 MSPERSQEESPKGDTERTERKPM 0 | ||
0 | 0 VKDAFKDISIYFTKEEWTEMGDWEKTRYRNVKRNYKTLITI 1 | ||
2 | 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPWMAFRGEQSKHQK 0 | ||
0 | 0 GMPKASFNNESSLKELSGTQNLLNTSGSEQAQKPVSPPGEASTSGQHSTLKI 1 | ||
2 | 2 ELRRKETEGKTYSLRERKGHAYKEVSEPQDDDYL 1 | ||
2 | 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDKEAANNGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 YVNCAWDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMPGR 1 | ||
2 | 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARHLLQAENPCPGDQNQEQQYSDPDCCNDKTKGQEIKERSKLLNKRTWQREISRAFSSSAKGQMGSSRVGERMMEEESGTGQKVNPGNTGKLFVGVGISRIAK | ||
VKYGECGQGFSDKSDVITHQRTHTGGRS | |||
YICRESGRGFTQKSGLLSHQRTHTGEKP | |||
YVCRECGWGFSQKSNLLRHQRTHTGEKP | |||
YVCRECGRGFSRKSVLLIHQRTHTGEKP | |||
<font color = red>*</font><font color = magenta>v</font>CRKDE | |||
> | >PRDM7_nomLeu Nomascus leucogenys (gibbon) ADFV01125891 pseu gas8+ synteny implied by non-coding | ||
0 0 | 0 0 | ||
0 1 | 0 1 | ||
2 | 2 1 | ||
2 0 | 2 IKSPWMAVRVEQSKHQK 0 | ||
0 1 | 0 GMPKASFNNESGLKELSGSQnLLNTSG<font color =red>*</font>EQARKPVSPPGEASTSGQHSRQKL 1 | ||
2 | 2 ELRRKETEGKMYSL<font color =red>*</font>ERKGHAYKEVSEPQDDDYL 1 | ||
2 | 2 YCEMCQNFFTDSCAAHGPPTFVKDSAVDKGHPNHSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKS<font color =red>*</font>ANWMK 2 | ||
1 | 1 YVNCARDHEEQNLVAFQYHRQIFYRTCQVIRPGCEPLVWYGDEYGQELGIKWGSKWKKELTAER 1 | ||
2 | 2 EPKPEIHPCPSCCLVFTSQKFLSQHVECNHSSQNFPGPSARKLLQRENPCPGDQNQEQQYSDSRSCNDKTKGQEIKERSKL<font color = magenta>n</font>KRIWQRKISRAFSSLPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK | ||
VKYGECGQGFSDKSDVIAHQGTHTGGKS | |||
<font color =red>*</font>ICRECGWGFSQESHLLIHQRTHTGEKL | |||
YVCRECGQGFSQKSDLLSHQRTHTGEKP | |||
YVRRECGRGFSQKSNLLSHQRTHTEEKP | |||
YVCRECGWGFSQKSHLLIHQRTHTGKKP | |||
<font color =red>*</font><font color = magenta>v</font>CRKDE | |||
>PRDM7_macMul Macaca mulatta (rhesus) pseu GAS8+ stop codon exon 5 in 6 of 6 trace reads, frameshift in exon 10 | |||
0 0 | |||
0 1 | |||
2 1 | |||
2 VKPPWMAFRVEQSKHQK 0 | |||
0 EMPKTSFNNESSLKELSGTPNLLSTSDSE<font color =red>*</font>AQKPASPPGEASTSGQHSRLKL 1 | |||
2 ELRRKETEGKMYSLRERKRHAYKEASELQHDDYL 1 | |||
2 YCEMCQNFFIDSCAAHGPPTFVKDNAVNKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPCEGRITEDKEAANSGYSWL 0 | |||
0 ITKGRNCYEYVDGKDKSWAKWMR 2 | |||
1 1 | |||
2 EPKPEIYPCPSCCLAFSSQKFLS<font color = magenta>q</font>HVERNHSSQNFPGPSARKLLQSENPCPGDQNQEQQYSDPSSCNDKTKGQEIKERSKLLNKRTWQREILRAFTSPPKGQMGSSRVGERMMEEEFRTGQKANPGNTGKLFVGVEISRIAK | |||
VKYGECGQGFSGKSDVITHQRTHTEGKP | |||
YVCRGCGRRFSQKSSLLRHQRTHTGEKP | |||
<font color =red>*</font><font color = magenta>v</font>CKKNE | |||
> | >PRDM7_macFas Macaca fascicularis (crab-eating macaque) CAEC01352986 CAEC01352983 4 frameshifts 1 stop codon | ||
0 0 | 0 0 | ||
0 1 | 0 1 | ||
2 | 2 1 | ||
2 0 | 2 0 | ||
0 | 0 EMPKTSFNNESSLKELSGTPNLLSTSDSE<font color = red>*</font>AQKPASPPGEASTSGQHSRLKL 1 | ||
2 | 2 ELRRKETEGKMYSLRERKRHAYKEVSELQHDDYL 1 | ||
2 | 2 YCEMCQNFFIDSCAAHGPPTFVKDNAVNKGHPNRSALSLPPGG<font color = magenta>l</font>RIGPSGIPQAGLGVWNEASDLPLGLHFGPCEGRITEDKEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 1 | ||
2 | 2 EPKPEIYPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQSENPCPGDQNQEQQYSDPSSCNDKTKGQEIKE<font color = magenta>r</font>SKLLNKRTWQREILRAFTSPPKGQMGSSRVGERMMEE<font color = magenta>e</font>FRTGQK<font color = magenta>a</font>NPGNTGKLFVGVEISRIAK | ||
VKYGECGQGFSGKSDVITHQRTHTEGKP | |||
YVCRGCGRRFSQKSSLLRHQRTHTG<font color = magenta>e</font>KP | |||
<font color = red>*</font><font color = magenta>v</font>CKKNE | |||
> | >PRDM7_papHam Papio hamadryas (baboon) genome Prim pseu 2 gas8+ contigs scattered | ||
0 | 0 MSPERSQEESPEEDTERTEWKPM 0 | ||
0 | 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 | ||
2 | 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPWMAVRVEQSKHQK 0 | ||
0 | 0 GMPKASFNNESSLKEVSGMANLLNTSGSEQAQKPVSPPGEARTSGQHSRLKL 1 | ||
2 | 2 ELRRKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 | ||
2 | 2 YCEMCQNFFIDSCAAHGPPTFIKDSAVEKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITQDEEAANNGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | ||
2 | 2 EPKPEIYPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQSENPCPGDQNQEQQYSDPSSCNDKTKGQEIKERSKLLNKRTRQRQILRAFTSPPKGQMGSSRVGERMMKEEFRTGQKANPGNTGKLFVGVEISRIAK | ||
VKYGECGQGFSDKSDVVIHQRTHTREKP | |||
YVYRgCGQGFSIKSNLLRHQRIHTGEKP | |||
> | >PRDM7_calJac Callithrix jacchus (marmoset) genome Prim gene 12 GAS8+ chr20 one frameshift in repeat area chr20 terminus | ||
0 | 0 MSPERSQEESPEGDTGRTEQKPM 0 | ||
0 | 0 VKDAFKDISMYFSKEEWAEMGDWEKTRYRNMKRNYNALITI 1 | ||
2 | 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPGMAFRVGQSKHQK 0 | ||
0 | 0 GMPKASFGNESSLKKLSGTANVLNTSGPEQAQKPVSPPGEASTSGQHSRLKL 1 | ||
2 | 2 ELRRKDTEEKMYSLRERKGLAYKEVSEPQDDDYL 1 | ||
2 | 2 yCEICQNFFIDSCAAHGPPTFVKDSAVDKGHPNHAALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRVTEDEEAASSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | ||
2 | 2 ESKPEIHPCPSCCLAFSSQKFLSHHVERNHSSQNFPGTSTRKLLQPENPCPGKQKEEQQYFDPCNSNDKTKGQETKERSKLLNIRTWQREMARAFSNPPKGQMGSSRVEERMMEEESRTGQKVNPVDTGKLFVGVGISRIAK | ||
AKYGECGQGFSDMSDVTGHQRTHTGEKP | |||
YVCRECGRGFSQKSALLSHQRTHTGEKP | |||
YVCRECGRGFSQKSHLLSHQRTHTGEK<font color=magenta>p</font> | |||
YVCTECGRGFSQKSVLLSHQRTHTGEKP | |||
YVCTECGRGFSRKSNLLSHQRTHTGEKP | |||
YVCRECGRGFSRKSALLSHQRTHTGEKP | |||
YVCRKCGRGFSQKSNLLSHQGTHTGEKP | |||
YVCTECGRGFSQKSHLLSHQRTHTGEKP | |||
YVCRKCGRGFSQKSNLLSHQRTHTGEKP | |||
YVCRECGRGFSFKSALLRHQRTHTGEKP | |||
YVCRECGRGFSRKSHLLSHQGTHIGEKP | |||
YVCRECGRGFSRKSNLLSHQRIHTGEKP | |||
YVRREDE | |||
>PRDM7_saiBol Saimiri boliviensis (squirrel_monkey) AGCE01149118/AGCE01147692/AGCE01012341/AGCE01145199 fragments no synteny but not in cadherin 10/12 complex | |||
MSPERSQEESP GDTGRTEQKPM | |||
VKDAFKDVAIYFSKEEWAEMGDWEKTRCRNVQRNYNALITI | |||
GLRATQPAFMCHRRQASKLQVDDTEDSDEEWTPRRQ | |||
RPVSPPGEASTSGQHSRVKP | |||
YVNCARDDEEQNLVAFQYHRQIFYRTCQVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR | |||
EPKPEIHPCPSCCLAFSSQKFLSRHVERNHSSQNFPGTSTRKLLQPENPCPGKQKEEQRYFDPCNSNDKTKGQEIKERSKLLNTRTWQREIARAFSYPPKGQMGSSRVEERMMEGESRTGQKVKPVDTGKLFVGVGISRIAK | |||
ANYGECGQGFSGMSDVTAQQRIHTGEKP | |||
YVCRECGRGFGHKSTLLSHQRTHTGEKP Y | |||
> | >PRDM7_tarSyr Tarsius syrichta (tarsier) ti|1493610848 ABRT011082008 ti|1633244849 ABRT010499286 ABRT010929713 ti|1646504284 pseu gas8? double frameshift in exon 5 single trace ti|1623660607 | ||
0 | 0 MSPDRSPEDSPEGDTGRTECKSA 0 | ||
0 | 0 VKDAFKDISIYFTKEEWAEMGDWEKIRYRNVKRNYNTLIAI 1 | ||
2 | 2 GLRAPRPAFMCHRKRAIKPLVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPWMAFRVEQNKHQK 0 | ||
0 | 0 GMPRAPLSIVSSLKELSEMANLL<font color=magenta>n</font>TSDSEQAWKPVSPS<font color=magenta>r</font>EASTSEQHSRKKI 1 | ||
2 | 2 EFRKKEIEVNMYSLRERKDCAYKEVNEPQDDDYL 1 | ||
2 | 2 YCEQCQNFFIDSCATHGIPTFINDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASELPLGLHFGPYEGQITDDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDKSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRIIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 | ||
2 | 2 ETKPKILPCSSCSLALSSQKFFSQHVKCNHPPQIFPGTSARKYVQPENPCPEDLNQEQQQSDPHSWNDKSKCQEAKERSKPMHKRTQQRESSRSLSNPPTEQTESSREKERMMKEEPSTSQKVHLGDTGYEFHHIGASAAR | ||
> | >PRDM7_micMur Microcebus murinus (lemur) ABDC01433247 ABDC01371462 gas8+ weak coverage last exon has two frameshifts | ||
0 | 0 MSPNKSQEESPEVDAGRTGWKPT 0 | ||
0 | 0 DKDAFKDISIYFSKEEWAQMGDWEKIRYRNVKRNYNVLITI 1 | ||
2 | 2 GLRAPRPAFMCHRRQAIKHQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 0 | ||
0 | 0 1 | ||
2 | 2 SEPQDDDYL 1 | ||
2 | 2 YCEKCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLKIRPSGIPQAGLGVWNEASELPLGLHFGPYEGQVTEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDDSWANWMR 2 | ||
1 | 1 YVNCARDEEEQNLVAFQYHRQIFYRTCQVIRPGCELLVWYGDEYGQELGIKWGSKWKEELTIRQ 1 | ||
2 | 2 EPKPEIHPCPSCSLAFSSQKFLSQHVKHTHSSQISPRTSGRKHLQPENPCPGDQNQEQQHSDPHSCNDKAKDQEVKERPKPFHKKTQQRGISRAFSSPPKGKMGSCREGKRIMEEEPRTGQKVGPGDTDKLCAAGGISRISR | ||
VKYGDSGQSFSDKSNVIIHQRTHTGEKP | |||
YVCRECGRGFSQKSDLLKHQRTHTGEKP | |||
YVCRECGRGFSQKSHLLRHQRTHTGEKP | |||
YVCRECGRGFSQKSDLLIHQRTHTGEKP | |||
YVCRECGRGFSCKSHLLIHQRTHTGEKP | |||
YVCRECGRGFSCKSSLLIHQRTHTGEKP | |||
YVCR<font color = magenta>e</font>CG<font color = magenta>r</font>GFSRKSDLLIHQRTHTGEKP | |||
CVCRKGE | |||
> | >PRDM7a_otoGar Otolemur garnettii (galago) AAQR03189271 (43912 bp) adjacent ARFGEF1 not GAS8 | ||
0 | 0 MSPNRSQEESPE 0 | ||
0 | 0 VRGAFKDTYKYFSMEELAEMGDWEKIHHGNVERNYDVLIDR 1 | ||
2 | 2 GLRAPQPAFMGHRRQAIKYQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 GKHSLMAFRMQPRKRQK 0 | ||
0 | 0 GMPRAPLSHDSILQDLSGPANSLNISDSEQHQNCVSLPGEANASGQNSRRKS 1 | ||
2 | 2 ALRRKEIEARSYNLRERTDRSYEEVSEPQNDDYL 1 | ||
2 | 2 YCEMCQDYFIDRCDVHGPPTFVKDIAVDKGHPNRAALTLPPGLSIRQSGIPQAGDGVWNEACELPLGLHFGPYEGQVLEDEEAARSGYAWK 0 | ||
0 | 0 ITKGRNCYEYVDGKDQSQGNWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHSQIFYRTCRVIRPGCELLVWYGTEYGKKLGIMWTSKRKKELTGQ 1 | ||
2 | 2 DPKPENHPCPSCSLAFSSQKSLSQHVEGTHSSQIFPGTSVRKHCRPEHLYPGDQNQEQQLSDPHDQNDKTKGQEMNEISKTSQEKTQQSSISGISSHTPEGQMGNSRDSERMVEPGQNMGPGETGKLCVKVEISRIVK | ||
VANGQCGQEFSQTSNLHTHQRTHTGEKP | |||
YVCSQCGHGFRYKSNLLTHQRIHTGEKP | |||
YICTECGQQFRQTSNLLAHQRIHTGEKP | |||
YVCSDCGKGFSQKSNLRTHQKTHTGEKA | |||
YVCSECGKGFTRKENLLIHHRTHTGEKP | |||
YICSDCGKRFSQKSNFLTHQKTHTGEKA | |||
CVCRECGKGFSRKATLLIHQRTHKGE<font color=magenta>k</font>P | |||
YVYRSCGQIFIHKSNLNRREKTHTGEKS | |||
> | >PRDM7b_otoGar Otolemur garnettii (galago) AAQR03144890 (36951 bp) adjacent GAS8+ pseudogene | ||
0 0 | 0 0 | ||
0 1 | 0 1 | ||
2 | 2 GLRAPRPAFMCHRRQATKYKVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VKHPWMAFRMEQSKRQK 0 | ||
0 | 0 ILKKCMLSFNMHLKELSGPASLPNISGSEQHQKHMSSPREASTSGQHSGRKS 1 | ||
2 | 2 DLRIKEIEVRMYSLRERKGHAYKEVSEPQDDDYL 1 | ||
2 | 2 yCEKCQNFFIDNCAVHGPPTFVKDTAVEKGHPNRSVLSLPSGLGIRTSGIPQAGFGVWNEASDLQLGLHFGPYEGQVTEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDESQGNWMR 2 | ||
1 | 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGQ 1 | ||
2 | 2 EPKPEIHPCPSCSLAFSTQKFLSQHVERTHPSQISQGTSGRKNLRPQTPCPRDENQEQQHSDPNSRNDKTKGQEVKEMSKTSHKKTQQSRISRIFSCPPKGQMGSSREGERMIEEEPRPDQKVGPGDTEKFCVAIGISGIV | ||
KVKNRECVQSFSNKSNLRHQRTHTGEKP | |||
YMCRDCGRGFSHKSSLFRHQRTHTGEKP | |||
YVCRDCGRGFSLKANLLTHQRTHTGEKP | |||
YVCRDCGQGFSQKAHLLRHQRTHTGEKP | |||
YMCRDCGQGFSRKAYLLTHQRTHTGEKP | |||
YVCRDCGQGFSQKAHLLTHQRTHTGEKP | |||
YVCRDCGRGFSHKSSLFRHQRTHTGEKP | |||
YICRDCG<font color=red>*</font>SFRDRSNLLRHQRTHTGEKL | |||
YVCRECGQGFNLKVTLLTHQRTHTGEKP | |||
YVCRDLG<font color=red>*</font>SFHNRSNLLTH<font color=magenta>q</font>RTHIGEKP | |||
YVCRDFGRGFSQKAHLLTHQR | |||
> | >PRDM7_tupBel Tupaia belangeri (tree_shrew) AAPY01316756 ti|1061183379 ti|1061388949 ti|1076935836 AAPY01523531 ti|1074533669 AAPY01523530 no synteny QVTSDKTYFSGQMEDKSGHHTPP* runout | ||
0 | 0 MRRYKSPEESPEGDAGRTEWKPT 0 | ||
0 | 0 VKDAFKDISVYFSKEEWAQMGEWEKIRYRNVKRNYTTLIAI 1 | ||
2 | 2 GLRAPRPAFMCHRKLAVKPHMDDAEDSDEEWTPRQQ 1 | ||
2 0 | 2 0 | ||
0 1 | 0 1 | ||
2 | 2 GKMYSLRERKCGTYKEVHEPQDDDYL 1 | ||
2 | 2 yCEKCQNFFIDSCSAHGPPIFVKDSAVDKGSLNRSVLSLPPGLRIAPSGIPEAGLGVWNAATDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDESCANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGEEYGQELGIKWGSKWKKSLWQGE 1 | ||
2 | 2 EPRPEIHPCLSCSLAFSSQKFLNQHVEHNHSCQRVPETSARKHLHPEHTHPGDEIQEQHHSDPQHCSDKADYEKVKERSKPSQERTRQRTMSRTFCSPCKGQMENAREGERMTGQRTDQKVGAEDTGKLLVRVGSSRIAG | ||
VKYGECGQGLINKSNASRHQRTHTGEKP | |||
YGCRECGRGFSQQSDLIRHQRTHTGEKP | |||
YLCGECGRGFSLQSSLIRHQRTHTGEKP | |||
YLCGECGRGFSRQSHLIIHQRTHTGEKP | |||
YVCRECGRGFSLQSNLIIHQRTHTGEKP | |||
YGCRECGRGFSQQSSLIRHQRTHTGEKP | |||
YLCGECGRGFSRQSHLIIHQRTHTGEKP | |||
YVCRECGRGFSLQSNLIIHQRTHTGEKP | |||
YGCRECGRGFSQQSSLIRHQRTHTGEKP | |||
YVCRECGRGFSRHSSLIIHQRTHTGEKP | |||
YLCGECGRGFSRQSHLIIHQRTHTGEKP | |||
YVCRECGRGFSQQPQLIIHQRTHTGEKP | |||
YVCRECGRGFRCQSHLIIHQRTHTGEKP | |||
YVCRECGRGFSQQPHLIIHQRTHTGEKP | |||
<font color=red>*</font>VCRKGE | |||
> | >PRDM7_oryCun Oryctolagus cuniculus (rabbit) genome Glir gene 8 other Un0161 exon 2 ttt to tt restores frame; ZNF717+ DCAF4+ YAP1+ PRDM9- qTer | ||
0 MSAAAPAEPSPGADAGQARGKPE 0 | |||
0 VQDAFRDISIYFSKEEWAEMGEWEKIRYRNVKRNYCALVAI 1 | |||
2 GLRAPRPAFMCHRRLAVRARADDTEDSDEEWTPRQQ 1 | |||
2 VKPPWMAFRTEHSKHQK 0 | |||
0 GMPRLPVNNESSLKELSGTANLLKTTGSEEDQKPSFPPKETRTSGQHSTRKL 1 | |||
2 GLRRKNIEVKMYSFRKRKSQAYKECSEPQDDDYL 1 | |||
2 YCEKCQNFFLDSCAVHGPPIFVKDSAVDKGHPNRSVLSLPPGLRIGPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEEEEAANSGYSWL 0 | |||
0 ITKGRNCYEYVDGKDRSWANWMR 2 | |||
1 YVNCARNDEEQNLVAFQYHKQIFYRTCQVIKPGCELLVWYGDEYGQELGIKWGSKWKEELTAGR 1 | |||
2 EPKPEIHPCPSCSLAFSSHKFLSQHMERSHSSQIFPGAPARNHLQPANPCPGKEHQKLSDPQSWNDKNEGQDVKEKSRFSSKRTRQKAISRSFSSLPKGQVETSREGERMIEEEPRIGQELNPEDTGKSSVGAGLSRIAG | |||
VKYRDCRQGLSDKSHLINGQRAHTGEKP | |||
YACRECERGFTVKSNLISHQRTHTGEKP | |||
YACRECGRGFTVKSALTTHQRTHTGEKP | |||
YACRECGRGFTVKSHLISHQRTHTGEKP | |||
YACRECGRGFTVKSALITHQRTHTGEKP | |||
YACRECGQGFTVKSNLISHQRTHTGEKP | |||
YACRECGRGFTQKSHLINHLRAHTGEKP | |||
YACRECGRGFTVKSDLISHQRTHTGEKP | |||
YACRVDE | |||
>PRDM7_ochPri Ochotona princeps (pika) ti|1534455888 AAYZ01312269 AAYZ01242582 no synteny | |||
0 0 | 0 0 | ||
0 | 0 VPDAFRDICVYFSRAEWAEMSESEKLRYRDVKRNYSALRAI 1 | ||
2 | 2 GFRAPRPAFMCRRRLAGRAREEDTDDSDEECTLRPQ 1 | ||
2 | 2 VKPPWMAFRTEHSKHQK 0 | ||
0 | 0 GIPKVPTHHESSVTEAPRTAPFLRPAGSQQGRKPAFPPEEASASGQHSTRRL 1 | ||
2 | 2 EGRRRKTDLKIYSLRKRKSRTYKECSEPQDDDYL 1 | ||
2 | 2 YCEMCQNFFIESCAVHGSPTFVKDNPVGKGHPHRSVLSLPSGLRIGPSGIPEAGLGVWNETTDLPLGLHFGPYEGQVTEEEEATNSGYSWL 0 | ||
0 | 0 ITKGRNRYEYVDGKDPSQANWMR 2 | ||
1 | 1 YVNCARNDEEQNLVAFQYHRQIFYRTCRAVRQGCELLVWYGDEYGQELGIKWGSKWKEELTAGR 1 | ||
2 | 2 EPKPEIHPCPSCSLAFSSHKFLSRHTERSHSSQVFPKTSPRRLQPVNPCRGTEHREPLDPHSWNDTAEGQEVIENSSSVSTRTRQRQISAAFCSLRKGQVEASREGERKAEESPRINQEVNAQDTAKSSVRAGLPSRVT | ||
VKCGDCRQDL | |||
> | >PRDM7_ratNor Rattus norvegicus (rat) P0C6Y7 Glir gene 10 PDCD2 chr1 FM103467 single transcript from body fat | ||
0 | 0 MNTNKPEENSTEGDAGKLEWKPK 0 | ||
0 | 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 | ||
2 | 2 GLRAPRPAFMCYQRQAIKPQINDNEDSDEEWTPKQQ 1 | ||
2 | 2 VSSPWVPFRVKHSKQQK 0 | ||
0 | 0 ETPRMPLSDKSSVKEVFGIENLLNTSGSEHAQKPVCSPEEGNTSGQHFGKKL 1 | ||
2 | 2 KLRRKNVEVNRYRLRERKDLAYEEVSEPQDDDYL 1 | ||
2 | 2 YCEKCQNFFIDSCPNHGPPVFVKDSVVDRGHPNHSVLSLPPGLRIGPSGIPEAGLGVWNEASDLPVGLHFGPYKGQITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGQDESQANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGRELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 | ||
2 | 2 ELRTEIHPCFLCSLAFSSQKFLTQHVEWNHRTEIFPGASARINPKPGDPCPDQLQEHFDSQNKNDKASNEVKRKSKPRHKWTRQRISTAFSSTLKEQMRSEESKRTVEEELRTGQTTNIEDTAKSFIASETS | ||
RIERQCGQCFSDKSNVSEHQRTHTGEKP | |||
YICRECGRGFSQKSDLIKHQRTHTEEKP | |||
YICRECGRGFTQKSDLIKHQRTHTEEKP | |||
YICRECGRGFTQKSDLIKHQRTHTGEKP | |||
YICRECGRGFTQKSDLIKHQRTHTEEKP | |||
YICRECGRGFTQKSSLIRHQRTHTGEKP | |||
YICRECGLGFTQKSNLIRHLRTHTGEKP | |||
YICRECGLGFTRKSNLIQHQRTHTGEKP | |||
YICRECGQGLTWKSSLIQHQRTHTGEKP | |||
YICRECGRGFTWKSSLIQHQRTHTVEK | |||
> | >PRDM7_musMus Mus musculus (mouse) genomic strain C57BL/6J 12 repeats 12 PDCD2 chr17 CN723438 eight transcripts, four from retina | ||
0 | 0 MNTNKLEENSPEEDTGKFEWKPK 0 | ||
0 | 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 | ||
2 | 2 GLRAPRPAFMCYQRQAMKPQINDSEDSDEEWTPKQQ 1 | ||
2 | 2 VSPPWVPFRVKHSKQQK 0 | ||
0 | 0 ESSRMPFSGESNVKEGSGIENLLNTSGSEHVQKPVSSLEEGNTSGQHSGKKL 1 | ||
2 | 2 KLRKKNVEVKMYRLRERKGLAYEEVSEPQDDDYL 1 | ||
2 | 2 YCEKCQNFFIDSCPNHGPPLFVKDSMVDRGHPNHSVLSLPPGLRISPSGIPEAGLGVWNEASDLPVGLHFGPYEGQITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGQDESQANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 | ||
2 | 2 ELRTEIHPCLLCSLAFSSQKFLTQHMEWNHRTEIFPGTSARINPKPGDPCSDQLQEQHVDSQNKNDKASNEVKRKSKPRQRISTTFPSTLKEQMRSEESKRTVEELRTGQTTNTEDTVKSFIASEIS | ||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTQNSHLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSNLIKHQRTHTGEKP | |||
YVCRECGWGFTQKSDLIQHQRTHTREK | |||
> | >PRDM7_musMol Mus molossinus (wild_mouse) GU216230 Glir gene 11 noDet full length deposit | ||
0 | 0 MNTNKLEENSPEEDTGKFEWKPK 0 | ||
0 | 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 | ||
2 | 2 GLRAPRPAFMCYQRQAMKPQINDSEDSDEEWTPKQQ 1 | ||
2 | 2 VSPPWVPFRVKHSKQQK 0 | ||
0 | 0 ESSRMPFSGESNVKEGSGIENLLNTSGSEHVQKPVSSLEEGNTSGQHSGKKL 1 | ||
2 | 2 KLRKKNVEVKMYRLRERKGLAYKEVSEPQDDDYL 1 | ||
2 | 2 YCEKCQNFFIDSCPNHGPPLFVKDSMVDRGHPNHSVLSLPPGLRISPSGIPEAGLGVWNEASDLPVGLHFGPYEGQITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGQDESQANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 | ||
2 | 2 ELRTEIHPCLLCSLAFSSQKFLTQHMEWNHRTEIFPGTSARINPKPGDPCSDQLQEQHVDSQNKNDKASNEVKRKSKPRQRISTTFPSTLKEQMRSEESKRTVEELRTGQTTNTEDTVKSFIASEIS | ||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTEKSSLIKHQRTHTGEKP | |||
YVCRECGWGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSSLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGWGFTQKSNLIKHQRTHTGEKP | |||
YVCRECGWGFTQKSDLIQHQRTHTREK | |||
>PRDM7_criGri Cricetulus griseus (hamster) AFTD01086355 no out-of-frame continuation, no synteny information in contig, no PRDM9 | |||
0 MSCTRNTNKQEGNSPAGDAERLEWKPK 0 | |||
0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 | |||
2 GLKAPRPAFMCYQRQAFKPQMDDSEDSDEEWTPKQQ 1 | |||
2 GSPPWVPFRVKHTKKQK 0 | |||
0 ETQRIPLNKESNVKEVSGSENLLSTSGSEHVQKTVFSPGEGNASGQHTGQKP 1 | |||
2 ELRRKNVEVKMYSLRERKDLAYEEVNEPQDDDYL 1 | |||
2 YCEKCQNFFINSCPSHGPPIFVKDSMVDRGHPNCSVLSLPPGLRIGPSGIPEAGLGVWNEASDLPVGLHFGPYKGQITDDEEAANSGYSWL 0 | |||
0 ITKGRNCYEYVDGQEESQANWMR 2 | |||
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVINPGCELLVWYGDEYGQELGIQWGRKNKKGFATGR 1 | |||
2 ELRTEIHPCLLCSLAFSSPKFLSQHVQWNHRTQIFPGASSTINSKPGDPHPDQLQEQQHFNSHNKNDKARSLEVKGKSKPMHKWTRQISTAFPSTLKGHMRSEENKKTMEVLRTGQKTNTEDTIKSFIGSEIS | |||
RIERKCGQYFSDKSNVNEHQRTHTGEKP | |||
YVCRECGRGFTQKSHLIRHQRTHTGEKP | |||
YVCRECGRGFTQKSNLIRHQRTHTGERP CVCLFKKDKKASVNKTTPQQSQKDKCSL* 0 | |||
> | >PRDM7_dipOrd Dipodomys ordii (kangaroo_rat) genome Glir gene -- noDet dubious fragment, no orthologous terminal exon | ||
0 | 0 0 | ||
0 | 0 1 | ||
2 | 2 GLKAPRPVFMCHRRQAIKPQVDDTDDSDEEWTPGRQ 1 | ||
2 | 2 0 | ||
0 | 0 1 | ||
2 | 2 elRTKEVKMRMYSLRERKSYAYEEISEPQDDDYL 1 | ||
2 | 2 yCEQCQNFFINSCTVHGPPIFVRDNVVDKGHYDRSVLSLPPGLRIRQSSIPEAGLGVWNEESDLPLGLHFGPYEGQITEDEDAANSGYSWM 0 | ||
0 | 0 ITKGRNCYVYVDGKDKSQANWMR 2 | ||
1 | 1 YVNCARYDEEQNLVAFQYHRQIFYRTCRVIKAGCELLVWYGDEYGQELGIKWGSKWKRELTAgr 1 | ||
2 | 2 | ||
> | >PRDM7_speTri Spermophilus tridecemlineatus (squirrel) AAQQ01308561 Glir gene -- noDet plus exon by exon traces | ||
0 | 0 0 | ||
0 | 0 1 | ||
2 | 2 GFRAPRPAFMCHQRQTIKLQMDDTEDSDEEWTPRQQ 1 | ||
2 | 2 0 | ||
0 | 0 LKPEVLLSNESSLKELSGTANLLNTSGSEQVQKPVSPLREASASRQHSRRKL 1 | ||
2 | 2 ELRTKEVEVKMYSLRERKGHAYKEVSEPQDDDYL 1 | ||
2 | 2 yCDKCQNFFMDSCPVHGPPTFIKDSVVNKDHSNHSTLSLPLGLRIGPSSIPEAGLGVWNEATDLPLGLHFGPYRGQITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDESQANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELSAGR 1 | ||
2 | 2 EPKPEIHPCPSCSLAFSSQKFLSQHVDRSHPSQIFPGTSMRKKLIPGDSSPRDQLQEQQHPDPHGWNDKARGQEVQGSLKPTHKGTRQRGISSPPKGQMGRSEESERMMEDDLKADQEINPEDTDKILVGVEMSRI | ||
>PRDM7_bosTau Bos taurus (cow) pseudogene on chr18 main Gas8 URAH synteny block not qTer not old, splice sites preserved | |||
0 MSPNRSPEESIEGDTGRTEWKPT 0 | |||
0 AKDAFKDISIYFCKEEWAQMG*WekA*YRNVKRNYEALITL 1 | |||
>PRDM7L1_bosTau Bos taurus (cattle) chrX:85801132 active gene 21.3 repeats formerly PRDM9b_bosTau, adjacent URAH pseudogene | |||
0 MSPNRSPENSTEGDAGRTEWKPM 0 | |||
0 AKDAFKDISIYFTKEEWAEMGEWEKIQYRNVKRNYEALIAI 1 | |||
2 GFRATQPGFMHHGRQVLKSQVDDTEDSDEEWTPRQQ 1 | |||
2 GKPSGMAFRGEPSKHPK 0 | |||
0 RLSRGPLNKVSSLKKLPGAAKLLKKSGSKQAQKPVPPPREARTPGKHPRHKV 1 | |||
2 ELRRKETEVKRYSVRERKGHVYQEVSEPQDDDYL 1 | |||
2 YCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQIIYNEEDSNSGYCWL 0 | |||
0 VTKGRNSYEYVDGKDTSLANWMR 2 | |||
1 YVNCARDDEEQNLVALQYHGQIFYRTCRVVRPGCELLVWYGDEYGEELGIKQDKRGKSKLSAQR 1 | |||
2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNQQQRYSDPHSLSDKPEGREPKERPHPLLKGPKLCIRPKRISTASSYPPKGQMGGSEVHERMTEEPSTSQKLNPEDTGKLFMEAGVSGIVR | |||
VNYGDHEQGSKDRSSLITHEKIHTGEKP | |||
YVCKECGKSFNGRSDLTKHKRTHTGEKP | |||
YACGECGRSFSFKKNLITHKRTHTREKP | |||
YVCRECGRSFNEKSRLTIHKRTHTGEKP | |||
YVCGDCGQSFSLKSVLITHQRTHTGEKP | |||
YVCGECGRSFNEKSRLTIHKRTHTGEKP | |||
YVCGDCGQSFSLKSVLITHQRTHTGEKP | |||
YVCGECGQSFNEKSRLTIHKRTHTGEKP | |||
YACGDCGQSFSLKSVLITHQRTHTGEKP | |||
YVCGECGQSFNEKSRLTIHKRTHTGEKP | |||
YVCGDCGQSFSLKSVLITHQRTHTGEKP | |||
YVCGDCGQSFSFKSVLITHQRTHTGEKP | |||
YACGECGRSFSGKSNLTKHKRTHTGEKP | |||
YVCGECGRSFSEKSNLTKHKRTHTGEKP | |||
YVCGDCGQSFSFKSVLITHQRTHTGEKP | |||
YVCGECGRSFSEKSNLTKHKRTHTGEKP | |||
YACGECGRSFSFKKNLITHQRTHTGEKP | |||
YVCRECGRSFSEKSRLTIHKRTHTGEKP | |||
YVCGDCGQSFSLKSVLITHQRTHTGEKP | |||
YVCRECGRSFSVMSNLIRHQRTHTGEKP | |||
YVCRECGRSFRVKSNLVRHQRTHTGEKP | |||
YVCMECE* 0 | |||
> | >PRDM7L2_bosTau Bos taurus (cattle) appears intact KRAB SSXRD SET C2H2 | ||
0 | 0 MSPNRSPENSTEGDAGRTEWKPK 0 | ||
0 | 0 AKDAFKDISIYFTKEEWAEMGEWEKIRYRNVKRNYEALIAI 1 | ||
2 | 2 GFRATQPGFMHHRRQVLKPQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 GKPSGMAFRGERSKHQK 0 | ||
0 | 0 RLSRGPLNKVSSLKKLPGAAKLLKKSGSKQAQKPVPPPREARTPGKHPRHKV 1 | ||
2 | 2 ELRRKETKVKRYSVRERKGHVYQEVSEPQDDDYL 1 | ||
2 | 2 YCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQIIYNEEDSHSGYCWL 0 | ||
0 | 0 VTKGRNSYEYVDGKDTSLANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVALQYHGQIFYRTCRVVRPGCELLVWYGDEYGEELGIKQDKRGKSKLSAQR 1 | ||
2 | 2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNEQQRYSDPHSLSDKPEGREPKERPHPLLKGPKLCIRLKRISTASSYPPKGQMGGSEVHERMTEEPSTSQKLNPEDTGKLFMEAGVSGIVR | ||
VKYGEHEQDSKDKSSLITHEKIHTGEKP | |||
YVCTECGKSFNWKSDLTKHKRTHSEEKP | |||
YACGECGRSFSFKKNLIIHQRTHTGEKP | |||
YVCGECGRSFSEKSNLTKHKRTHTGEKP | |||
YACGECGQSFSFKKNLITHQRTHTGEKP | |||
YVCGECGRSFSEKSRLTTHKRTHTGEKP | |||
YVCGDCGQSFSLKSVLITHQRTHTGEKP | |||
YVCRECGRSFSVISNLIRHQRTHTGEKP | |||
YVCRECEQSFREKSNLVRHQRTHTGEKP | |||
YVCMECE* 0 | |||
> | >PRDM7L3_bosTau Bos taurus (cattle) appears intact KRAB SSXRD SET C2H2 probable assembly stutter also has URAHps | ||
0 | 0 MSPNRSPENSTEGDAGRTEWKPK 0 | ||
0 | 0 AKDAFKDISIYFTKEEWAEMGEWEKIRYRNVKRNYEALIAI 1 | ||
2 | 2 GFRATQPGFMHHRRQVLKPQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 GKPSGMAFRGERSKHQK 0 | ||
0 | 0 RLSRGPLNKVSSLKKLPGAAKLLKKSGSKQAQKPVPPPREARTPGKHPRHKV 1 | ||
2 | 2 ELRRKETKVKRYSVRERKGHVYQEVSEPQDDDYL 1 | ||
2 | 2 YCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQIIYNEEDSHSGYCWL 0 | ||
0 | 0 VTKGRNSYEYVDGKDTSLANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVALQYHGQIFYRTCRVVRPGCELLVWYGDEYGEELGIKQDKRGKSKLSAQR 1 | ||
2 | 2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNEQQRYSDPHSLSDKPEGREPKERPHPLLKGPKLCIRLKRISTASSYPPKGQMGGSEVHERMTEEPSTSQKLNPEDTGKLFMEAGVSGIVR | ||
VKYGEHEQDSKDKSSLITHEKIHTGEKP | |||
YVCTECGKSFNWKSDLTKHKRTHSEEKP | |||
YACGECGRSFSFKKNLIIHQRTHTGEKP | |||
YVCGECGRSFSEKSNLTKHKRTHTGEKP | |||
YACGECGQSFSFKKNLITHQRTHTGEKP | |||
YVCGECGRSFSEKSRLTTHKRTHTGEKP | |||
YVCGDCGQSFSLKSVLITHQRTHTGEKP | |||
YVCRECGRSFSVISNLIRHQRTHTGEKP | |||
YVCRECEQSFREKSNLVRHQRTHTGEKP | |||
YVCMECE* 0 | |||
> | >PRDM7L4_bosTau Bos taurus (cattle) chr21 distal frameshift, stop codon, deletion: pseudogene adjacent to URAHL3 pseudogene | ||
0 | 0 MNPYRSPEESTEGDAGRTEWRWR 0 | ||
0 | 0 1 | ||
2 | 2 1 | ||
2 | 2 GKPSWMTFRVKHSKHQK 0 | ||
0 | 0 gMSRALVSNKSSLKELPGASKLLKTSGPKQAQIQCPLPEKQVPLNSTLDKKW 1 | ||
2 | 2 GPIQKETEVKMYSLRERASHVYQEVSEPQDDDYL 1 | ||
2 | 2 YCEKCENF IKSCAVHGLPTFVKDCAVEKGHVNRLALSVPAGLSIRPSGIPEAVLGVWNEVSDLPLALHFGSYKGQIIDDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDTSWAKWMR 2 | ||
1 | 1 YVNCARDDKEQNLVAFLSHRQIFYQTCPVVRPGCELLVWYGDKYSQELSIKCGSRWKSELMASR 1 | ||
2 | 2 EPKPKIYPCASCSLAFSSQKFLSQHAEHNHPSQILLRTSARDRLQTKDSCPGNQNHQQQYSDPHSWRDKPEDREVKERPQPLLQSVRLRRVSRASSYSPKGQMGDTWVSERMMQEPSTGQKVNTEDTGKLCMGAGVLTIIR | ||
VKSveCGQDSKDRSSLITHQRIHTGEKP | |||
YACRECGRNFSEKSPLIRPQRTHIGEKP | |||
FVCRECE*GFSH IKHQRTHTGEKP | |||
YVCRECEQSFSEKSTLIRHQTTHTGEKP | |||
YVCGEVE* | |||
> | >PRDM7L5_bosTau Bos taurus (cattle) intact somewhat diverged chr1:161387010- no URAH | ||
0 | 0 MRPNTSPEESTERDAGRTEWKPT 0 | ||
0 | 0 AKDAFKDISVYFSKEEWEEMGEWEKIRYRNVKRNYEALIAI 1 | ||
2 | 2 GFRATRPAFMHHRRQVIKLQADDTEDSDEEWTPRQQ 1 | ||
2 | 2 GKLSSMAFRVEHNKHQN 0 | ||
0 | 0 TMSRAPLSKEFSLKELPGAAKLLKTSGSKQAQKLVPPPGKARTPGQHPRQKV 1 | ||
2 | 2 ELRRKETEVKRYSLRERKGHVYQEVSEPQDDDYL 1 | ||
2 | 2 YCEECQDFFIDSCAAHGPPIFVKDCAVEKGHANRSALTLPPGLSIRESSIPEAGLGVWNEVSDLPLGLHFGPYEGQITDDEEAANSGYSWL 0 | ||
0 | 0 ITKRRNCYEYVDGKDTSLANWMR 2 | ||
1 | 1 RYVNCARDDEEQNLVALQYHGQIFYRTCQVVRPGCELLVWYGDEYGQDLGIKRESSRKSELAGPR 1 | ||
1 ESKPKIHPCASCSLAFSSQKFLSQHVQHNHPSQTLLRPSARDYLQPEDPCPGSQNQQQRYSDPHSPSDKPEGREVKDRPQPLLKSIRLKRISRASSYSPRGQMGASGVHERITEEPSTSQKPNPEDTGKLFMGAGVSGIIK | |||
VKYGECGQGSKDRSSLITNQRTHTGEKP | |||
YVCGECGQSFNQKSTLITHQRTHTGEKP | |||
YVCGECGRSFNQKSTLITHQRTHTGEKP | |||
YVCGECGRSFSQKSTLIKHQRTHTGEKP | |||
YVCGECGQSFNQKSTLITHQRTHTGEKP | |||
YVCGECGQSFNQKSTLITHQRTHTGEKP | |||
YVCGECGRSFSRKSTLITHQRTHRGEAL CLQGV* | |||
>PRDM7_oviAri Ovis aries (sheep) original PRDM7 pseudogene adjacent to GAS8 URAH on chr14, first two exons only (as in cow) | |||
0 MSPNRFP*ESTGGDPGRTEWKPM 0 | |||
0 AKDAFKDISIYFSKEEWAEMR*W 1 | |||
> | >PRDM7L1_oviAri Ovis aries (sheep) active gene ChrX:5148004 frameshift exon 2 may be read error; translocated with URAH2 pseudogene to ChrX both minus strand | ||
0 | 0 MSPNRSPEKSTEGDAGRTEWKPM 0 | ||
0 | 0 AKDAFKDISIYFTKEEWAEMGEWekVQYRNVKRNYKALIAI 1 | ||
2 | 2 GFRATQPAFMHHHRQVIKPQVDDTEDSEEEWTPRQQ 1 | ||
2 | 2 GKPSGMAFRGEPSKHQK 0 | ||
0 | 0 RLSRGPLNKVSSLKKLPGAAKLLKKTGSKQAQKPVPPPREARTPGQHPRHKV 1 | ||
2 | 2 ELRRKETEVKRYSLRERKGHVYQEVSELQDDDYL 1 | ||
2 | 2 yCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQVIYNEEASHSGYSWL 0 | ||
0 | 0 VTKGRNSYEYVDGKDTSLANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVALQYHGQIFYRTCQVVRPGCELLVWYGDEYGEELGIKQDSRGKSKLSAQR 1 | ||
2 | 2 EPKPKIHPCASCSLSFSSQKFLSQHVQRSHPSQILLRPSPRDHLQPEDPCPGKQNQQQRYSDPHSPSDKPEGQEPKERPHPLLKGPKLCIRLKRISTASSYTPKGQMGGSEVHEKMTEEPSTSQKLNPENTGKLFMEAGVSGIVR | ||
VKYGEHEQGSKDKSSLITHERIHTGEKP | |||
YVCKECGKSFNGRSNLTRHKRTHTGEKP | |||
YVCRECGQSFSLKSILITHQRTHTGEKP | |||
YVCGECGQSFSEKSNLTRHKRTHTGEKP | |||
YVCRECGQSFSLKSILITHQRTHTGEKP | |||
YVCRECGRSFSVKSNLTRHKMTHTGEKP | |||
YVCGECGQSFSQKPHLIKHQRTHTGEKP | |||
YVCRECGRSFSAMSNLIRHQRTHTGEKP | |||
YVCRECGRSFSAMSNLIRHQRTHTGEKP | |||
YVCRECGRQVSSHTRGHTQGRSPMFAGSVGEASV* | |||
>PRDM7L2_oviAri Ovis aries (sheep) OARX:67135299 not tandem, two frameshifts and internal stop: pseudogene | |||
0 0 | |||
0 1 | |||
2 1 | |||
2 0 | |||
0 GMSRGPLSKVSSLKKLPGTTKLLKTSGSKQAQKPVPSSREARTSG*HTRQKV 1 | |||
2 eLRRKETEVKRYSL*ERKGHVYQVVSEPQNDNYL 1 67134368 | |||
2 ycEECQNFFMNSCAAQVPPTFVKDSAVGKGHANCSALTLPPGLSIRLSGIPEARLGVWNEVSDLPLGLHFGpyEGQITDDEEAAHSGDSWL 0 | |||
0 ITKGRNSFEYVDGKDMSLANWMR 2 67133322 | |||
1 CVKCTQDNKEQNLVALQYHRQIFYRICQVVRPGcqLLVWYGDEYAVELGIKWDNRGKSEFTARR | |||
2 ELKPKIHPCASCSPAFSSQKFLSQYVQPNHPSQILLRPSARDHLQPEDPCPGNQNEQQ*YSDPHSPSDKPEGCKAKERPPWLLKSMSVRISMASSYSPKGQMRGSETHYRMTEEPSTSQKLNPEDIGKLFMGTGVSGIIK | |||
IKYEECGQVSKDRSSLITHEGTHTREQS | |||
YVCRECGQSFSVKSSLIRLQRTHTGEKP | |||
YT ESVGKASVRTPISSHTRGHTQERSP | |||
MVAGRVGKASVRTQFSSDNRGHTQGRSP | |||
MFSGSVGKASVRSPSSSHTRGHAPE | |||
>PRDM7L3_oviAri Ovis aries (sheep) chrX:67,417,035+ 2 internal stops: pseudogene | |||
0 MS*NRSPQERTEGDAGRTEWKLM 0 | |||
0 ANGAFKNISIYFSKEEWAEMGEWEKI*YGNVKRNCEALIAI 1 | |||
2 GFRATRPAFMHHRRQVIKPQGNDTEDSDEEWTPWQQ 1 | |||
2 0 | |||
0 GKSRGPLSKASSLKKLPGAAKLLKKSGSKWAQEPVKPPRETRTPGQHSRQKV 1 | |||
2 ELGRKETDMKRYSLRERKGHVYQEVSEPQDDDYL 1 | |||
2 yCQECQNFFINSCDAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRLSGIPEAGLGVWNEASHLPLGLHFGPYEGQITDDKEAVNSGYSWL 0 | |||
0 ITKGRKSYEYVDGKDTSLANWMR 2 | |||
1 YVNCARHYEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGEKLGIRCESRGKSMLAAGR 1 | |||
2 * 0 | |||
>PRDM7L4_oviAri Ovis aries (sheep) OAR18:28239884-one frameshift, one stop codon pseudogene paired with URAH3 pseudogene | |||
0 MNPYISPEESTEGDAERTEWKPM 0 | |||
0 AKDAFKDISIYFSKEECAEMGEWGKICYRNAKRNCEALITI 1 | |||
2 GLRATRLAFMHHCRQVIKPQVDDIEDSDEEWTPRQQ 1 | |||
2 0 | |||
0 EGMSKALVSNKSSLKEMPGASKLLKTRGPKQAQIPVPAPREPSTSEQHPRQKV 1 | |||
2 1 | |||
2 CCHGLPTLVKDCAVEKGHANHSALSLSPGSSIRPSGIPEAGLGVWNKVSDLLLGLHFGSYVGQITDDEEAAKSGYSWL 0 | |||
0 ITKGRNCYEYVDGKQRSWANWIr 2 | |||
1 YVNGAQD KEQNLVAFLTHRQIFY*TCRVVRPGCELLVWYRDTYSQELSIKCGSRWKSELTASR 1 | |||
2 EPKPKIHPCVSCSLsfSSQKFLRQHVERNHPSQILLRTSARDRLQTKDSCPGNQNHEQQYCDPHSWSDKPEDGEVRERPQPLLKSIRLRRVSRASSYSPKGQMGDSWVSEKMMEEPSTGQKLNTEGTGKLCMGAGVLRIIR | |||
VKHGECGQGSKDRSSLITHQRIHNGEKP | |||
YVCRECGQSFSEKSILIRHQRTHTGEKP | |||
FVCRECERGFSQKSYLIRHQKTHTGEKS | |||
YVCREVE* | |||
> | >PRDM7L5_oviAri Ovis aries (sheep) OAR5:40765355+ 1 frameshift and 4 internal stops: pseudogene | ||
0 | 0 MSSNRSLKERTEGDARRTEWKPMV 0 | ||
0 | 0 AKDAFKDISI*FSKEEWAEMGE*EKI*YRNVKRNYEALITTI 1 | ||
2 | 2 GLRAP*PPFMYHRRQVIKPQVDDIEDSDEEWTPRQQ 1 | ||
2 | 2 0 | ||
0 | 0 GMSRVPLSN ESMKELLGAAKLLT SGSKQAQKPVPPPREASTSEQHPRKKV 1 | ||
2 | 2 ELRRKETEMKIYSLQKRKGHMYQEVSDPQDDNYL 1 | ||
2 | 2 0 | ||
0 | 2 ycEKCQNF INSCAAHGPPTFVKDCVVEKGHASCSALtlSPGLSIRPSGIPEAGLRVWNEASDLPLGLHFGPYKGQITDDEEVANSRYFWL 0 | ||
0 2 | |||
2 | 0 ITKGRNCYEYVDGRD | ||
1 2 | |||
1 YVNCAQDDEEQNLVAFQYHRQIFS*TCWVVRPGCELLVWYRDEYGQELSIK*GSRHKSELTVRR 1 | |||
2 PMCSCSLAFSSQKFLSQHVKCNHPSQILLKTSARDRLQPEDPCPGNPNQQQQYSDLHSWSDKPESRESKEKPQPLLKS----IRLRRISRASSYSSRGQMGGFRVHKRMREEPSTGKEVSPEDAGKLFMGEGVSRIM | |||
VKYGDCG*GSKDRSSLMTHQRTHTGENP | |||
> | >PRDM9d_munMun Muntiacus muntjak (muntjac) AC216498 Laur gene 4 noDet frameshift exon 9 no syntenic loci; identities: 92%b 89%a 90%c | ||
0 | 0 MRPNRSQEESTEGNAGRTERKPT 0 | ||
0 | 0 GKDAFKDISVYFSKEEWEEMGEWEKIRYRNMKRNYEALIAI 1 | ||
2 | 2 GFRATQPTFMHHRRQVIKSQVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 GKPSSMAFRVEHSKNQK 0 | ||
0 | 0 RMSRAPLSNESGLKELPGAAKSLKTSDSKQARNPVPHHRKARTPGQLPRQKV 1 | ||
2 | 2 ELRRKETGVKRYSLRERKGHVYQEVSEPQDDDYL 1 | ||
2 | 2 YCEECQNFFINSCAAHGPpTFVKDCAVEKGHANRSALTLPHGLSIRLSGIPDAGLGVWNKVSDLALGLHFGPYKGQITDNEEAANSGYAWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDTSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGQDFGIKRNSRGKSELAAGR 1 | ||
2 | 2 EPKPKIHPCASCSLTFSSQKFLSQHIQCSHPPQTLLRPSERDLLQPEDPCPGNQNQQQRYSDPHSPSDKPEGHEAKDRPQPLLKSIRLKRISRASSCSPRGQMGGSGVHERMTEEPSTSQKLNPGDTGTLLTGAGVSGIMK | ||
VKYGECGQGSKDRSSLSTHERTHTGEKP | |||
YVCRECGQSFSGKPVLIRHQRTHTGEKP | |||
YVCMECGRSFSAKSVLMTHHRTHTGEKP | |||
YICRECGQSFSQKIHLIRHQRIHTGE.P | |||
SVFRECE | |||
>PRDM9c_munMun Muntiacus muntjak (muntjac) AC154919 Laur gene 15 noDet no syntenic loci AC204173 99% identical | |||
0 MRPNRSPEESTEGDAGRTEQKPT 0 | |||
0 AKDAFKDISVYFSKEEWEEMGDWEKIRYRNMKRNYEVLIAI 1 | |||
2 GFRATRPDFMHHRRQVIKPQVDDTEDSDEEWAPRQQ 1 | |||
2 GKPSSVAFRVEHSKHQK 0 | |||
0 RMSRAPLSNESGLKELPGAAKPLKTSGSKQAQNPVPHHRKARTPGQLPRQKV 1 | |||
2 ELRRKETGVKRYSLRERKGHVYQEVSKPQDDDYL 1 | |||
2 YCEKCQNFFIDSCAAHGPPTFVKDCAVEKGHANRSLLTLPPGLSIRLSGIPDAGLGVWNEASDLPLGLHFGPYEGQITDDEEAANSGYAWL 0 | |||
> | 0 ITKGRDCYQYVDGKDTSWANWMR 2 | ||
0 | 1 YVNCARDDEEQNLVAFQYHGQIFYQTCQVVRPGCELLVWCGDEYGQDLGIKRNSRGKSELVAGR 1 | ||
0 | 2 EPKPKIHPCASCSLAFSSQKFLSQHIQRSHPSQTLLRPSERDLLQPEDPCPGNQNQRFSDPHRPSDRPQPLLKSIRLKRISRASSYSPRGQMGGSGVHELMTEEPSTSHKLNPEDTGTLLMGAGVSGIMR | ||
2 | VTYGECGQGSKDRSSLTTHERTYTGEKP | ||
2 | YVCGECGRSFCQKAHLITHQRTHTGEKP | ||
0 | YVCRECGQSFSRNSLLIRHQRIHTGEKP | ||
2 | YVCGECGRSFRDKSNLISHRRTHTGEKP | ||
2 | YVCGECGQSFSDKSNLIRHQRTHAGEKP | ||
0 | YVCGECGRSFNRKSHLITHQRTHTGEKP | ||
1 | YACRECGQSFSQKSILITHQRTHTGEKP | ||
2 | YACRECG.SFSQKSILITHQRTHTGEKP | ||
YVCGECGRSFSQKSLLITHQRTHTGEKP | |||
YVCMECGRSFSQKTHLITHQRTHTGEKP | |||
YVCGECGRSFSQKSLLITHQRTHTGEKP | |||
YVCGECGRSFSQKSLLITHQRTHTGEKP | |||
YICMECGRSFSQKTHLITHQRTHTGEKP | |||
YVCGKCGQSFSDKSNLISHKRTHTGEKP | |||
YVCRECGRSFNRKSLLITHQRTHT.E.P | |||
YVCRECE | |||
> | >PRDM9b_munMun Muntiacus muntjak (muntjac) AC218859 Laur gene 13 noDet no syntenic loci | ||
0 | 0 MRPNTSPEESTEGDAGRTERKPT 0 | ||
0 | 0 AKDAFKDISVYFSKEEWEEMGDWEKSRYRNMKRNYEVLIAI 1 | ||
2 | 2 GFRATRPDFMHHRRQVIKPQVDDTEDSDEEWAPRQQ 1 | ||
2 | 2 GKPSSMAFRVEHSKHQK 0 | ||
0 | 0 RMSRAPLSNESGLKELPGAAKPLKTSGSKQAQNPVPHHRKARTPGQLPRQKV 1 | ||
2 | 2 ELRRKETGVKRYSLRERKGHVYQEVSKPQDDDYL 1 | ||
2 | 2 YCEECQNFFIDSCAAHGPPTFVKDCAVEKGHANRSALTLPPGLSIRLSGIPDAGLGVWNETSDLPLGLHFGPYEGQITDDEEAANSGYAWL 0 | ||
0 | 0 ITKGRNCYQYVDGKDTSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVIRPGCELLVWYGDEYGQDLGIKRNSRGKSELATGR 1 | ||
2 | 2 EPKPKIHPCASCSLAFSSQKFLSQHIQRSHPSQTLLRPSERDLLQPEDPCPGSQNQRYSDPHSPSDKPEGQEAKDRPQQLLKSIRLKRISRASSYSPGGQMGGSGVHERMTEEPSTSQKLNPEDTGTLLTGAGVSGIMR | ||
VTYGECWKGSKDRSSLTTHERTHTGEKP | |||
YVCGECGQSFHHGSVLIRHQRTHTGEKP | |||
YVCGECGRSFSQKSVLIRHQRTHTGEKP | |||
YVCGECGRSFSQKSVLIRHQRTHTGEKP | |||
YVCGECGRSFSQKAHLITHQRTHTGEKP | |||
YVCGECGRSFSQKTHLISHKRTHTGEKP | |||
YVCGECGRSFCQKSALIRHQRAHTGEKP | |||
YVCGECGRSFIQKSDFIRHQRTHTGEKP | |||
YVCRECGQSYSDKTVLITHERTHTGEKP | |||
YVCGECGRSYSDKTVLITHERTHTGEKP | |||
YVCGECGRSFLWKSALIRHQRTHTGEKP | |||
YACGDCGRSFNQKSNFIRHQRTHTGEKP | |||
YVCGECWRSFSQKSSSSDTRGHTQGRRP | |||
VCRECG SFSQKSHLISHQRTHTEEKP | |||
YVCRECE | |||
> | >PRDM9a_munMun Muntiacus muntjak (muntjac) AC225653 Laur gene 7 noDet unordered contigs htgs; no synteny tag stop instead of aag K | ||
0 0 | 0 MRPNRSPEESTEGDAGRTEQKPT 0 | ||
0 AKDAFKDISVYFSKEEWEEMGEWEKIRYRNVKRNYEALIAI 1 | |||
2 GFRATRPDFMHHCRQVIKPQVDDTEDSDEEWTPRQQ 1 | |||
2 GKPSSMAFRVKHSKHQK 0 | |||
0 GMSRAPLIKESSLKELLGAAKLMKTSGSKQAQNPVPHPRKARTPGQHPRQKV 1 | |||
2 ELTRKETGVKRYSLRERKGHVYQEVSEPQDDDYL 1 | |||
2 YCEECQNFFIDSCAAHGLPTFVKDCAVEKGHANRSALTLPPGLSIRLSGIPDAGLGVWNEESDLPLGLHFGPYEGQITDDEEAANSGYAWL 0 | |||
0 ITKGRNCYQYVDGKDTSWANWMR 2 | |||
1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVIRPGCELLVWYGDEYGQDLGIKRNSRGKSELAAGR 1 | |||
2 EPKPKIHPCASCSLAFTSQKFLSQHIQRSHPAQTLLRPSERNLLQPEHPCPGSQNQRYSDPHSLSDKPEGQEAKDRPQPLLKSIRLKRISRASSYSPGGQMGGSGVHERMKDEPSTSQKLNPEDTGTLLTGAGVSGIMR | |||
VTYGECGKGSKDRSSLTTHERTHTGEKP | |||
YACRECGRSFRQKSDFITHQRTHTGEKP | |||
YVCGQCGRSFGRKFALIRHQRIHTGEKP | |||
YVCRECGQSFSQKTHLSSHQRTHTGEKP | |||
YVCGECGRSFSQKSVLIRHQRTHTGEKP | |||
YVCQECGRSFSDKSNLISHKRTHMGEKP | |||
YVCRECGRSFIRKSVLIRHQRTHTGE.P | |||
YVCRECE | |||
>PRDM9a_odoVir Odocoileus virginianus (deer) AEGZ01043838/AEGZ01024932/AEGZ01044038/AEGY01012861/AEGZ01038568/AEGZ01003977/AEGY01011331/AEGZ01039403/AEGY01006151 possibly chimeric, frameshifts possibly errors in low coverage assembly | |||
0 MTVSRSLEESTGGDAGRTEWKPT 0 | |||
0 AFKDISKYFSKEEWARLGYSEKISYVYMKRNYETMTRL 1 | |||
2 GFRATRPAFMHHRRQVIKPQVDDTEDSDEEWTPRQQ 1 | |||
2 GKPSRMALREEHIKHQK 0 | |||
0 RMSRAPLSKESSLKELPGAAKSLKTSGSKQAQKPVPHPRKARTPGQHPRQKV 1 | |||
2 ELRRKETGVKRYSLRERKGHVYQEVSEPQDDDYL 1 | |||
2 yCEECQNFFIDSCAAHGPPTFVKDSVVKRGHANRSALTLPPGLSIRLSGIPDAGLGVWNEASDLPRGLHFGPYEGQITDDEEAANSGYAWL 0 | |||
0 ITKGRNCYEYVDGKDTSWANWMR 2 | |||
1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGQFLGIKRDSR<font color=magenta>g</font>KSKLAAGR 1 | |||
2 EPKPKIHPCASCSLAFSSQKFLSQHIQCSHPSQTPP<font color=magenta>r</font>PSERDLLQPEDPCPGNQNQRYSDPHSPSDKPEGQEAKDRPQPLLKSIRLKRISRASSYSPGGQMGGSGVHE | |||
>PRDM7_bosTau Bos taurus (cattle) genome Laur pseu -- GAS8+ missing C2H2 | |||
0 MSPNRSPEESIEGDTGRTEWKPT 0 | |||
0 AKDAFKDISIYFCKEEWAQMG WEKIRYRNVKRNYEALITL 1 | |||
2 1 | |||
2 0 | |||
0 1 | 0 1 | ||
2 1 | 2 1 | ||
2 | 2 0 | ||
0 | 0 2 | ||
1 1 | |||
2 | |||
1 | |||
2 | |||
> | >PRDM7_turTru Tursiops truncatus (dolphin) ABRN01441536 Laur gene 9 gas8+ no useful synteny | ||
0 | 0 MSTDRWPEDSTEGDAGRTAWKPT 0 | ||
0 | 0 VKDAFKDISIYFSKEEWTEMGEWEKIRYRNVKKNYEALVTL 1 | ||
2 | 2 GLRAPRPAFMCHRRQAIKAQVGDPEDSDEEWTPRQQ 1 | ||
2 | 2 VKPSWVAFRVEHSKHQK 0 | ||
0 | 0 AVPPVPLSNESSLKKLPGAAQLQKASGPAQAQSPAPPPGAASTSAWHTRQKL 1 | ||
2 | 2 ERRAKQIEVKMYSLRERKGHVYQEVSEPQDDDYL 1 | ||
2 | 2 yCEKCQNFFIDSCAAHGAPTFVKDSAVEKGHPNRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDTSWANWMR 2 | ||
1 | 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYSQELGIPWGSGWKSQLVaGR 1 | ||
2 | 2 DPKPKIQPCGSCSLAFSSQKILSQHVECSHPSQVLPRTSARDRVQPEDPCPGYQNRQQQYSDPHSWSNKPECQEVKERSKPLLKRIRLGRISRAFSSSPKGQMGSSRAHERMMEAGPSTGQKVNPEATGKLLIGAGVSRVVK | ||
VKYRSSGQGSKDRSSLTKHQRTHTGEKP | |||
YVCGECGRDFSLKSDLIRHQRTHTGEKP | |||
YVCGECGRDFSLKSGLISHQRTHTGEKP | |||
YVCGECGRDFSQKSGLIRHQRTHTGEKP | |||
YVCGECGRDFSLKSGLISHQRTHTGEKP | |||
YVCGECGRDFSQKSGLIRHQRTHTGEKP | |||
YVCGECGRDFSLKSGLITHQRTHTGEKP | |||
YVCGECGRDFSQKSNLITHQRTHTGEKP | |||
YVCGECGRDFSRKSSYI | |||
> | >PRDM7_susScr Sus scrofa (pig) FP476134 Laur gene 9 GAS8+ unordered HTGS not wgs misassembly or inversion; not in genome browser | ||
0 MRPDRRPEESPDPAAGSTERKAA 0 | |||
0 ATDAFKDISIYFSKEEWTEMGEWEKIRYRNVKRNYEALTTI 1 | |||
2 GLRAPRPAFMCHRRQAIKPQVDDTEDSDEEWTPRQQ 1 | |||
2 VKPCRVAFRVEHNKHQK 0 | |||
0 SDSRVPLSNKSSLKELLTTAEVPETSGSEQAQEPVSPPGEASTSRRRSGQEL 1 | |||
2 ARRRKDTEARMYSLRERKGHAYQEVGEPQDDDYL 1 | |||
2 yCEKCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALTLPPGLRIRPSGIPEAGLGVWNEAHDLPLGLHFGPYEGQVTEDEEAANSGYSWL 0 | |||
0 ITKGRNCYEYVDGKDKSWANWMR 2 | |||
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGI 1 | |||
2 EPKPKIHPCPSCSLAFSSQRFLSQHVERSHPSQSLPRASARRGLQPEGPCPDNQQQQQPYPDPHSWDGTSESQDVKEGSKPFLERRRLRKTSRASSYAPEGQMRSSRVRERMTEEEPSAGQKVNPEDTGTLFTVAGES | |||
GILRVENRGYGPDSGLTRHPRTHTGEKP | |||
HVCSECGRGFSVKSHLIRHQRTHTGEKP | |||
YVCRECGRGFSVKSHLIRHQRTHTGEKP | |||
YVCRECGRGFSVKSSLITHQRTHTGEKP | |||
YVCRECGRGFSVKSHLIRHQRTHTGEKP | |||
YVCRECGRGFSEKSSLVTHQRTHTGEKP | |||
FVCRECGRGFSVKSSLVTHQRTHTGEKP | |||
YVCRECGRGFSVKSNFITHQRTHTGEKP | |||
YVCRECGRGFSEKSSLVTHQRTHTGEKP | |||
YVCREGE | |||
>PRDM7_lamPac Lama pacos (llama) scaffolds traces | |||
0 0 | 0 0 | ||
0 | 0 TFKDISIYFSKEEWTEMGEWEKIRYRNVKRNYEALITI 1 | ||
2 | 2 GLRAPRPAFMCHRRKAIKPQVDDTEDSDEEWTPRQQ 1 | ||
2 0 | 2 0 | ||
0 | 0 GMPRGPLSNQSSLKELSGTAKPLKTSGSGQAQKPFPPLGEASTSGRHSRQKL 1 | ||
2 | 2 ELRRKESQVKMYSLRERKGHAYQEVSEPQDDDYL 1 | ||
2 | 2 0 | ||
0 | 0 ITKGRKCYEYVDGKDKYWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGEEYGQELGIKWGSKWKKSLWQGE 1 | ||
2 | 2 EPKIYLCPSCSLAFSSQKFLSQHVKHNHPSQILPRTAAGRHLEPEDPCPGNQNEQQQHSDQHSWNDKPEGQEAKERSKPFLKRIRLRRISGAFSYSHKGQMGNSRVHDRMIEEEPSTGQKVNPKDTGKLFTWAGVSRTVE | ||
VNYGEYGQGCKDTSHLTTHQRTHTGEKP | |||
YVCRECGRGFTRKSNLTIHQREHTTGEK | |||
>PRDM7_canFam Canis familiaris (dog) genome Laur pseu 5 GAS8+ frameshift fixed to 6 ZNF; synteny MNS1 K1F1B intervening CDH3 oddity | |||
> | |||
0 0 | 0 0 | ||
0 | 0 AFKDISKYFSKEEWAKLGYSDKITYVYMKRNYDTMTGL 1 | ||
2 | 2 GLRATLPAFMCPKKRAIKSK<font color=green> </font>RHDSDENENHRNQ 1 | ||
2 | 2 VKPSWVAFRMEQSKHQK 0 | ||
0 | 0 GIPRVPLSNKSSLKELSETAKLLNTSSPEQGQKSVSLPGKASTSGHHTRQKL 1 | ||
2 | 2 ELRRKDVEVKMYSLQERKGLAYQEVSEPQDDDYL 1 | ||
2 | 2 yCEK<font color=red>*</font>QTFFIDSCTVHGPPTFVKDSEVDKGQPNHSALTLPPGLRIRTSSIPQAGLGVWN<font color=red>*</font>ASDLPLGLHFGPYKGQITEDEEAANSGYSCL 0 | ||
0 | 0 ITKGRNCYEYVDG<font color=blue>KD</font>KSWANWMR 2 | ||
1 | 1 YMNCARDDEEQ<font color=magenta>s</font>LVAFQYHRQIFYR<font color=magenta>t</font>PGHQASCELLVWYGDEYSQELGIKWGSKWKSELTAGK 1 | ||
2 | 2 EPNPEIHPCPSCSL<font color=magenta>a</font>FSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR | ||
VKYRGCGRGFNDRSHLSRHQRTHTGENP | |||
YVCRECGRGFIHRTNLIIHQRTHTGEKP | |||
YVCRECG<font color=magenta>r</font>GFIQRSNLSIHQRTHTGEKP | |||
YVCRECGRGFTQRSTLNEHQRTHTEEKP | |||
YVCRECGRSFTRRSTLITHQRTHTGEKP | |||
YVCRECGRSFT | |||
>PRDM7_canLup Canis lupus (gray_wolf) JF750654 first frameshift: 1 bp del ggcctt to gcctt; second frameshift 1 bp del ggaca to ggacGa | |||
ePNPEIHPCPSCSL<font color=magenta>a</font>FSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR | |||
VKYRGCGRGFNDRSHLSRHQRTHTGENP | |||
YVCRECGRGFIHRTNLIIHQRTHTGEKP | |||
YVCRECG<font color=magenta>r</font>GFIQRSNLSIHQRTHTGEKP | |||
YVCRECGRGFTQRSTLNEHQRTHTEEKP | |||
YVCRECGRSFTRRSTLITHQRTHTGEKP | |||
YVCRECGRSFTKRS | |||
>PRDM7_canAur Canis aureus (golden_jackal) JF750659 | |||
EPNPEIHPCPSCSL<font color=magenta>a</font>FSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR | |||
VKYRGCGRGFNDRSHLSRHQRTHTGENP | |||
YVCRECGRGFIHRTNLIIHQRTHTGEKP | |||
YVCRECGRGFTQRSTLNEHQRTHTEEKP | |||
YVCRECGRSFTKRST | |||
> | >PRDM7_lycPic Lycaon pictus (painted_dog) F750657 | ||
EPNPEIHPCPSCSL<font color=magenta>a</font>FSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWSDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIM<font color=magenta>r</font> | |||
VKYRGCGRGFNDRSHLSRHQRTHTGENP | |||
YVCRECGRGFTHRTNLIIHQRTHTGEKP | |||
YVCRECG<font color=magenta>r</font>GFIQRSNLSIHQRTHTGEKP | |||
YVCRECGRGFTQRSTLNEHQRTHTEEKP | |||
YVCRECGRSFTRRSTLITHQRTHTGEKP | |||
YVCRECGRSFTKRST | |||
>PRDM7_canMes Canis mesomelas (black-backed_jackal) JF750658 | |||
EPNPEIHPCPSCSL<font color=magenta>a</font>FSSQKFLSQHLEHNHPSQILPQISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR | |||
VKYRGCGRGFNDRSHLSRHQRTHTGENP | |||
YVCRECGRDFTHRTNLIIHQRTHTGEKP | |||
YVCRECG<font color=magenta>r</font>GFIQRSNLSIHQRTHTGEKP | |||
YVCRECGRGFTQRSTLNEHQRTHTEEKP | |||
YVCRECGRSF<font color=magenta>t</font>RRSTLITHQRTHTGEKP | |||
YVCRECGRSFTKRST | |||
>PRDM7_speVen Speothos venaticus (bush_dog) JF750656 | |||
EPNPEIHPCPSCSL<font color=magenta>a</font>FSSQKFLSQHLEHNHPSQILP<font color=red>*</font>ISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKRIRQRRISRAFSTPCKGQTTCEGIVKEEPSASSQKLNPEDTGKLFKGVGMTRIIR | |||
VKYRGCGRGFNDRSHLSRHQRTHTGENP | |||
YVCREC<font color=magenta>g</font>RGFTHRTNLIIHQTTHTGEKP | |||
YVCRECG<font color=magenta>r</font>GFIQRSNLSIHQRTHTGEKP | |||
YVCRECGRSFT<font color=red>*</font>RSTFSThQRTH | |||
>PRDM7_vulVul Vulpes vulpes (red_fox) JF750655 more distal frameshif | |||
EPNPEIYPCPSCSLSFSSQKFLSQHLEHNHPSQILPRISIREHFQPKDPCPGCQNQQQQQHSDPQCWNDRAKGQEGKERFKPLP<font color=magenta>k</font>SIRQRKISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR | |||
VKYRGCGRGFNDRSHLSRHQRTHMGENP | |||
YVCRECGRGFTHRTNLIIHQRTHTGEKP | |||
YVSWECGRSFTRRSNLITHQRTHTGEKP | |||
YVCRECGRGFTKRSTLSTHQRTHL | |||
> | >PRDM7_neoVis Neovison vison (mink) JF288183 anomalous array, contig terminates | ||
0 | 0 0 | ||
0 | 0 1 | ||
2 | 2 1 | ||
2 | 2 VRPSWVAFRMEQSKHQK 0 | ||
0 | 0 GIPRAPLSNESSLKELSETAKLLNTSGSEQGQKPVSHPGEASTSGHHSLRKL 1 | ||
2 | 2 ELRRKDVEVKMYSLRERKSLAYQEVSEPQDDDYL 1 | ||
2 | 2 YCEKCQNFFIDSCAVHGPPTFVKDSAVDKGQPNRSDLTLPPGLRIRPSGIPQAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL | ||
0 | 0 ITKGRNCYEYVDGKDNSWANWMR 2 | ||
1 | 1 1 | ||
2 | 2 EPKPEIHPCPSCTLAFSSQKFLSQHLKCNHPSQILPRISAGEHFQPEDPCPGEQNHQQQQHSDPQSWNDKAKGQEVKESFKPLLESIRQRRNSRAFPTPCKGQTGYEGMVEEESSTGQKLNPEETEKLFMGVGMSRMIR | ||
VKYRGSGQGFDDRSHLSRHQRTHKEEKP | |||
SVGKELRREFIHKSVLVTHQRTHTEALP | |||
> | >PRDM7_musPut Mustela putorius (ferret) AEYP01035076 AEYP01035077 AEYP01035078:GAS8- HUIH+ CAD1L+ distal PRDM7+ | ||
0 | 0 MRPRTASESEQGLPGGPSTGSVSGPPEETPERDSGRTGRKPP 0 | ||
0 | 0 AQDAFKDISVYFSKEEWTEMGDWEKIRYRNVKRNYEALITI 1 | ||
2 | 2 GLRAPRPAFMCHRRQATIPRVDDTEDSDEEWTPRQQ 1 | ||
2 | 2 VRPSWVAFKMEQSKHQK 0 | ||
0 | 0 GVPRAPLSNESSLKELSETAKLLNTSGSEHDQKPVSHPGEASTSGHHSLRKL 1 | ||
2 | 2 ELRRKDVEVKMYSLRERKSLAYQEVSEPQDDDYL 1 | ||
2 | 2 YCEKCQNFFIDSCAVHGPPTFVKDSAVDKGQPNRSALTLPPGLRIRPSGIPQAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDNSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYRRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKSELTAEK 1 | ||
2 | 2 EPKPEIHPCPSCTLAFSSQKFLSQHLERNHPSQILPRISAGEHFQPEDPCPGEQNHQQQQHSDPQNWNDKAKGQDVKESFKPLLESIRQRKNSRAFPIPCEGQTGYEGIVEEEPSTGQKLNPEETGKLFMGVGMSRIIR | ||
VKYRGSGQGFDDRSHLSRHQRTHKEEKP | |||
SVGKEPRREFIHKSVLVTHQRTHTGEKP | |||
YVCRECGRGFTQRSHLIRHQRthtgEKP | |||
YVCRECGRGFTQRSNLITHHRTHTGEKP | |||
YVCRECGRGFTRRSNLIRHHRTHTGEKP | |||
YVCRECGRGFTWRSHLITHQRTHTGEKP | |||
YVCRECGRGFTWRSHLIRHQRTHTGEKP | |||
YVCRECGRGFTRRSNLITHQRTHTEALP INCISMTRGKM* | |||
> | >PRDM7_ailMel Ailuropoda melanoleuca (panda) GL193502 Laur gene 6 GAS8+ first three exons from different contig ACTA01106867 | ||
0 | 0 MGPLPASESEQSLPGGPSTMSLNTSPEETPERDSGRTGWKPT 0 | ||
0 | 0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYEALITI 1 | ||
2 | 2 GLRAPRPAFMCHRRQAIKPQVDDTEDSDEEWTPRRQ 1 | ||
2 VRPSWVAFRMEQSKHQR 0 | 2 VRPSWVAFRMEQSKHQR 0 | ||
0 | 0 GIPRAPLRNESSLKELSETAKLLNTSGSELGQKPVSLPGEASTSGHDSLQKL 1 | ||
2 | 2 GFRRKDVEVKMYSLRERKSLAYQEVSEPQDDDYL 1 | ||
2 | 2 yCEKCQNFFIDSCAVHGPPTFVKDSAVDKGQPNRSALTLPPGLRIRPSGIPQAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDNSWANWMR 2 | ||
1 | 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKSELAAGK 1 | ||
2 | 2 EPKPEIHPCPSCSLAFSSQKFLSQHLEHNHPSQILSRKSASEHFQQEDPCPGHQNQQQQQHSDPHRWNDKAKGQEVKERFKPLLKSIRQRRISRAFSSPCKGQTRSSTVCEGMVEEEPSAGQKLNPEETGKLFMGVGMSGIIR | ||
VKYRGCGRDFSDRSHQSGHQRRH QKKP | |||
SVCKK<font color=blue>V</font>KREFSHKSVLITHQRTHTGEKP | |||
YVCRECGRGFTQRSNLIRHQRTHTGEKP | |||
YVCRECGRGFTQRSNLIRHQRTHTGEKP | |||
YVCRECGRGFTQRSSLIRHQRTHTGEKP | |||
YVCRECGRGFTLRPNLIGHQRTHT<font color=blue>E</font>ALP INYISTTKEQM | |||
> | >PRDM7_felCat Felis catus (cat) genome Laur gene 11 GAS8+ two contigs GAS8 implied by downstream CAD1 | ||
0 | 0 MEPSPASESARGQPGGPGTTSPLRFPEQSAERGSRKARWKPT 0 | ||
0 | 0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYEALMTI 1 | ||
2 | 2 gLRAPRPAFMCHRRQAIKPQVDVTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPSWVASRVDQNKQHK 0 | ||
0 | 0 GTHRVPLSKESSLKDFSETAKLLNTSGSEQGQKPVSLPGEASTSGHHSRRKL 1 | ||
2 | 2 ELRRKEIGVKMYSLRERKGFAYQEVSEPQDDDYL 1 | ||
2 | 2 yCEKCQNFFIDSCAVHGPPTFVKDNAVGKGHPNRSALTLPPGLRIRPSSIPEAGLGVWNEASDLPLGTHFGPYEGQITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDNSWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKSELSTGK 1 | ||
2 | 2 EPQPDIHRCPSCSLAFSSQKFLSQHVECKHSSQSLPQISARKHFQPENPCPGDQNQQQQQHSDPHSWNDKAKCQEVKERSRPLLKSIKQRRISRAFSTPCKGQMGSSRVCEGMVEEGPSMGQNLNSEDTGKLFMGVGMSRIVR | ||
IKNRGCEQGFNDRSHFSRHQRTHKEEKP | |||
SVCNEFRRDFSHKSALITHQRTHTGEKP | |||
YVCRECGRGFTQRSNLFRHQRTHTGEKP | |||
YVCRECGRGFTQRSDLFTHQRTHTGEKP | |||
YVCRECGRGFTRRSNLFTHQRTHTGEKP | |||
YVCRECGRGFTRRSHLFTHQRTHTGEKP | |||
YVCRECGRGFTQRSNLFTHQRTHTGEKP | |||
YVCRECGRGFTQRSDLFRHQRTHTGEKP | |||
YVCRECGRGFTQRSHLFTHQRTHTGEKP | |||
YVCRECGRGFTQRSNLFRHQRTHTGEKP | |||
YVCRECGRGFTWRSNLFTHQRTHTGEKP | |||
YVCRKDGQGFTNKLHLSYQRT | |||
NVATTHSIPQL | |||
> | >PRDM7_equCab Equus caballus (horse) genome Laur gene 4 GAS8+ missing front exons, pre-terminal stop GAS8+- flanked right by EMR2- | ||
0 0 | 0 0 | ||
0 | 0 1 | ||
2 | 2 1 | ||
2 | 2 VKPSWVAFRVEQSKQQK 0 | ||
0 | 0 RMRTAPLSNESRLKELSGTAKLLKTSSSEQVQKPVSPLGEASSSEQHSRRKL 1 | ||
2 | 2 ELRRKEVGVKMYSLRERKGHAYQEVSEPQDDDYL 1 | ||
2 | 2 yCENCQNFFIDSCAAHGPPIFVKDSAVDKGHPNRSALTLPLGLRIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDISWANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYGQELGIKWGSKWKRELTAGR 1 | ||
2 | 2 EPKLEIHPCPSCSLAFSSQKFLSQHVERNHPSQILPGTSARNHLQPEDPSPGDQNQQQQHSDPHSWKDKAHSQEVKERSKPLLKKIRQRRIPRAFSYPPKGQMENFRMRERIMEEKPSIGRKVNPEDTGKLFLEMRMSRNVR | ||
VQYGGCGRGFNDRASLIKHQRTHTGEKP | |||
YVCRECEQGFTQKSSLIAHQRTHTGEKP | |||
YVCRECEQGFSEKSHLIRHQRTHTGEKP | |||
YVCRECEQGFSVKSNLIRHQRTHTGEKL | |||
<font color=red>*</font>FCREGK | |||
>PRDM7_pteVam Pteropus vampyrus (bat) ABRP01250178 Laur gene 7 GAS8+ 4 distal exons of GAS8+-; unique F sweep in zinc finger; 15 ZNF dotplot no CAD1 | |||
0 MRPDRSPEEAPEGDTRRTGCKPK 0 | |||
0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYDALQAI 1 | |||
2 GLRAPRPAFMCRRRQAIKPQVDDSEDSDEEWTPRQQ 1 | |||
2 0 | |||
0 AMPRVPLSNEPSLKELSVIANLLKASGSEQDQKPVFPPGKASASRQHSRQKL 1 | |||
2 GLRRKGVEVKMYSLRERTGRVYQEVSEPQDDDYL 1 | |||
2 yCEKCQNFFIDSCAAHGSPIFVKDSEVDIRHPNRSALTLPPGLRIGPSGIPEAGLGVWNEASDLPLGLLFGPYEGQVTEDEEAANSGYSWL 0 | |||
0 QGKGRNCYEYVDGKDESRANWMR 2 | |||
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKRELTAGR 1 | |||
2 EPKPAIHPCPSCSLAFSGQKFLSQHMKRSHPSQSLPGISARKHLQSKEPHPEDQSQQQHNDPRSWNDKAEGQEVKERSKPLLERNRQRKIFRAFSKPPKGQMGSPREYERMMEAEPSTSQKVNPENTGKSSVGVGASRIVI | |||
VKYGGCEHGFDDGSHLIMHQRTHSGEKP | |||
FVCRECERGFSKKSNLITHQRTHSGEKP | |||
> | FVCRECERGFTRKSSLITHQRTHSGEKP | ||
0 | FVCRECERGFTQKSHLITHQRTHSGEKP | ||
0 | FVCRECERGFSEKSSLIKHQRTHSGEKP | ||
2 | FVCRECERGFTRKSSLITHQRTHSGEKP | ||
2 | FVCRECERGFTQKSSLIKHQRTHSGEKP | ||
0 | FVCRECERGFTQKSSLIKHQRTHSGEKP | ||
2 | FVCRECERGFTQKSSLIKHQRTHSGEKP | ||
2 | FVCRECERGFTQKSSLITHQRTHSGEKP | ||
0 | FVCRECERGFTQKSHLITHQRTHSGEKP | ||
1 | FVCRECERGFSKKSNLITHQRTHSGEKP | ||
2 | FVCRECERGFTRKSLLITHQRTHSGEKP | ||
FVFRECERGFTQKSSLITHQRTHSGEKP | |||
FVCRECERGFTRKSYLITHQRTHSGEKP | |||
FVGRECE | |||
> | >PRDM9_pteVam Pteropus vampyrus (bat) ABRP01232219 Laur pseu 15 noDet frameshift ttt to tttt fixed in last zinc finger; no blastx synteny | ||
0 0 | 0 0 | ||
0 | 0 1 | ||
2 | 2 1 | ||
2 0 | 2 vQPSWVAFGVEQSKHQK 0 | ||
0 AMPRVPLSNESSLKELSVIANPLKASGSEQNQQPVFPPGKASASRQHSRRKL 1 | |||
2 eLRRKGVEVKMDSLRERMGRVYQEVSEPQDDDYL 1 | |||
2 yCEKCQNFFIDSCAAHGSPIFVKDSEVDIGHPNHSALTLPPGLRIGPSGIPEAGLGVWNEASNLPLGLLFGPYEGQVTEDEEAANSKYSwM 0 | |||
0 spKGETAEYV DGKDESRANWMR 2 | |||
1 YVNCARDDEDQNLVAFQFRRQIFYRTCRVIMPGCELLVWYGDEYGQGLGIKWGSKWKREFTAGR 1 | |||
2 EPKPEIHPCPSCSLAFSSRKFLSQHMKRSHPSQSLPGISARKHLQSKEPHPEDQSQQQQQQQHTDPCSWNDKAEGQEVKERSKPMLERNGQRKISRAFSKPPKGQMGSPRECERMMEAEPSTSQKVNPENTGKSSVGVGASRIVR | |||
VKYGGCGHGFDDGSHFIRHQRTHSGEKP | |||
FVCRECERGFNEKSSLTMHQRTHSGEKP | |||
FVCREC.EGFSVKSSLIRHQRTYSGEKP | |||
FVCRECEQGFNEKSSLTMHQRTHSGEKP | |||
FFCRECEGFSVK.SSLIRHQRTHSGQKP | |||
FVCRECKRGFTQKSHLITHQRTHSGEKP | |||
FCRECER.GFTQKSHLIKHQRTHSGEKP | |||
FVCRECA | |||
>PRDM7_myoLuc Myotis lucifugus (bat) AAPE02062260 Laur gene 6 gas8+ TGA stop codon; CpG hotspot for R CGA; SXXRD implies missing KRAB no CAD1 | |||
0 0 | |||
0 1 | 0 1 | ||
2 1 | |||
2 1 | |||
2 0 | 2 0 | ||
0 | 0 AKSRAPLSNESSLKELSGTANLLTTSGSEQTQKTVPPPGEASTSGQHPRSKL 1 | ||
2 | 2 dLRRKEIEVKMYSLRERKCRVYQEISEPQDDDYL 1 | ||
2 | 2 YCEKCQNFFIDSCAVHGPPTFVKDSAVDKGHANRSALTLPPGLRIGPSGIPEAGLGVWNEECDLPVGLHYGPYEGQITEDEAIANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGKDTSQANWMR 2 | ||
1 | 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVVRKGCELLVWYGEEYGQELGIKWGSKWKTEPVAGR 1 | ||
2 | 2 EPKPEIHPCPSCSVAFSSQTFLSQHGKRNHPSEILPGAPAGNHLQSEEPGPERQNQQQQQQTGPHGWNDKAEGQEVKGRSKPLLKRIRQRGTSRASFKPPNRHMGSSSERERIREEEPSTGQNVNHKNTGKLFVGVKRSKSVT | ||
IKHGGCGQGFNDGSHIDTHQRTHSGEKP | |||
YICRECGGFTHKSDL IRHQRTHSQENP | |||
YVCRECGRGFRDRSTLITHQRTHSGEKP | |||
YVCRECGRGLTEKSTLITHQRTHSGEKP | |||
YVCRECGRGFTRKSTLITHQRTHSGEKP | |||
YVCRECGRGSRVKSNLIRHQRTHSGEKS GVCIEGE | |||
> | >PRDM7_sorAra Sorex araneus (shrew) AALT01000095 Laur gene 8 noDet no useful synteny; upstream spectrin, IgG; GAS8 contig has no sign of pseudogene | ||
0 0 | 0 MSLNRPAEMNTQGKARKLMLKPM 0 | ||
0 | 0 SKDAFKDISMYFSKEEWAEMGDWEKIRHRNVKRNYEELISI 1 | ||
2 | 2 GLRAARPAFMSHRRQAIKTQLDDTEESDEEWTPNQQ 1 | ||
2 | 2 VKSLRVAFRAEQSKHQK 0 | ||
0 1 | 0 GRSRTPISNESSSKELSGTRTLLNTKCTKQAQKPLFPPGEASTSGHYSKPKL 1 | ||
2 | 2 ELRRKEPEVKMYSLRERKGRAYQEVSEPQDDDYL 1 | ||
2 | 2 YCENCQNFFINKCSAHGSPIFVKDNAVAKGHSNRSALTLPHGLRIGPSGIPEAGLGIWNEASDLPLGLHFGPYEGQITNDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNCYEYVDGVDESLANWMR 2 | ||
1 | 1 YVNCARDYEEQNLVAFQYHRQIFYRTCRIIKPGCELLVWYGDEYGQELGIKWGSKWKSELTADK 1 | ||
2 EPKPEIYPCPCCSLAFSNQKFLSRHVEHSHPSLILPGTSARTHPKSVNFCPGDQNQWQQHSDACNDKPDEPWNDKLENHKSKGRSKPLPKRMGQKRISTAFPNLRSSKMGSSNKHETIMDKINTGQKENPKDTYRVFAGIGMPRIIR | |||
DKHVTLRRSFTNRSSPLTHQRTHTGEKP | |||
YVCRECGRGFSQKSHLLTHQRTHTGEKP | |||
YVCRECGRGFTDRSSLLTHQRTHTGEKP | |||
YVCRECGRGFSLKSSLLRHQRTHTGEKP | |||
YVCRECGRGFSLKSSLLTHQRTHTGEKP | |||
YVCRECGRGFTDRSSLLTHQRTHTGEKP | |||
2 | YVCRECGRGFSLKSSLLTHQRTHTGEKP | ||
YVCRECGRGFSRKSSLLRHQRTHTGEKP | |||
YVCES | |||
>PRDM7_echEur Echinops europaeus (hedgehog) ti|970966337 | |||
epkpeihpcPSCSLAFSAQKFLNQHVKHSHPSQILPGTSTRKQPQVENPCLSNQNQQKQHSNFQNQHDSTESQEAIEKFKPLLKMIKQKTISNGFSKLPKEQIGSSREHEKTKEEESNSCQKMNPEDTSELLVGLGMSRIV | |||
DKYEGSGKNYYDMSHIITHQKTHTGEKP | |||
HVCKECGRGFSEKSSLIAHQRTHTGEKP | |||
HVCRECGRGFSEKSSLIAHQRAHTGEKP | |||
HVCRECGRGFSEKSGLITHQSTHTGEKP | |||
HVCRECG<font color=magenta>r</font>GFSAKSSLIAHKRTHTGEKA PCLQGSVGELQ | |||
> | >PRDM9a_loxAfr Loxodonta africana (elephant) genome Afro gene 12 noDet chr 153 novel synteny THEG+ MIER2+ PPAP2C PRDM9- ZNF699- | ||
0 0 | 0 MSPARAAKKNPRGDVGSAGRTPT 0 | ||
0 | 0 aKDTFRDISIYFSKEEWAEMGEWEKFRYRNVKRNYEALVTI 1 | ||
2 | 2 GLRAPRPAFMCHRRQAIKAQVDNTEDSDEEWTPRQQ 1 | ||
2 | 2 VKPPSVASRAEQSRHQK 0 | ||
0 1 | 0 GTPKALLGNESSLKEVSGTAILLNTTGSEQAQKPVSSPGEASTSDQPSRWKL 1 | ||
2 | 2 EPRRNEVEVKMYNLRERKGLEYQEVSEPQDDDYL 1 | ||
2 | 2 yCEKCQNFFIDTCAVHGAPMFVKDSPVDRGHPNHSALTLPPGLRIGPSSIPKAGLGVWNEASELPLGLHFGPYEGQVTEDKEAANSGYSWL 0 | ||
0 | 0 ITKGKNCYEYVDGKDESWANWMR 2 | ||
1 | 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRTIQPDCELLVWYGDEYGQELGIKWGSRWKKELTSGR 1 | ||
2 | 2 EPKPEIHPCPSCRLAFSSQKFLSQHMKHSHPSPPFPGTPERKYLQPEDPRPGGRRQQRSEQHMWSDKAEDPEAGDGSRLVFERTRRGCISKACSSLPKGQIGSSREGNRMMETKPSPGQKANPEDAEKLFLGVGTSRIAK | ||
VRCGECGQGFSQKSVLIRHQKTHSGEKP | |||
YVCGECGRGFSVKSVLIKHQRTHSGEKP | |||
YVCGECGRGFSVKSVLITHQRTHSGEKP | |||
YVCGECGRGFSVKSVLITHQRTHSGEKP | |||
YVCGECGRGFSQKSDLIKHQRTHSGEKP | |||
YSCRECGRGFSRKSVLITHQRTHSGEKP | |||
YVCGECGRGFSQKSNLITHQRTHSGEKP | |||
YVCGECGRGFSRKSVLITHQRTHSGEKP | |||
YVCGECGRGFSQKSNLITHQRTHSGEKP | |||
YVCGECGRGFSQKSDLITHQRTHSGEKP | |||
YVCRECGRGFSRKSNLITHQRTHSGEKP | |||
YVCRECRRGFSVKSALI GHGRRKCSKSAEPLHFPRVSRDQK | |||
> | >PRDM9b_loxAfr Loxodonta africana (elephant) genome Afro pseu 3 noDet approx seq after frameshift correction | ||
0 0 | 0 0 | ||
0 1 | 0 1 | ||
2 1 | 2 1 | ||
2 0 | 2 0 | ||
0 1 | 0 GTPKVLLSNESSLKEVSGTAILLSTMGSEQAQKPVSSPGEASTSDQPSRRKQ 1 | ||
2 | 2 EPRRKEVEVNMYSLRERKGLVYQEVGEPQDDDYL 1 | ||
2 | 2 yCEKCQNFFIHTCAVHGAPMFVKDSHVDRGHLNHSALTLPPGLRIGPSSIPEAGLRVR EVSEQLLGLHIGPYEGQVTEDkEAAHSGYSWL 0 | ||
2 | 0 ITKGRNCYKYVDGKDDPWANRMR 2 | ||
1 | 1 YVNCIQD KEQNLVAFQYHRQIFHWTCCTIRPGCELLVWYGDNYSQELGIKWGSR KKEL 1 | ||
2 | 2 EPKPEIHPCPSCPLAISSQKFLDQHTKHSHPSPPFPGTPERKHLQPEDPHPGGRRQQHSEQHLNDKAEDPETGDGSKPVFERARLVGGGAGGVSKVCSSLPKGQMGSSREGNRMMETEGQKVNPEDTEKLFLGVGISRLAK | ||
VRCGEYGQGFSQKSVLIRHQRTYSGEEH | |||
YVCGECGRGFSWKSQLTRHQRSHSWEKP | |||
YVCRECGGFSVKSTLI GTGEGNAATIHLHLPS | |||
>PRDM7_loxAfr Loxodonta africana (elephant) genome Afro pseu 5 GAS8+ scaffold_57 several frameshifts; ZNF540 opposite strand upstream of N-terminus | |||
> | 0 0 | ||
0 0 | 0 1 | ||
0 1 | 2 GLRASHPAFTCHCMQAIKAQMDDTEDSNEEQTPRQq 1 | ||
2 1 | 2 VRPSWVAFRMEQSKHQR 0 | ||
2 0 | 0 GMLRVPRSNESSLKNLSGTSIMLSRAGSEQAQKLVLPPGKASTSDEHSRQKP 1 | ||
0 1 | 2 EHRRKGVEVKMYSF ERKGLVYQEIS PQDDDYL 1 | ||
2 | 2 YCEKCQNFFIDTCESHGVPTFVKNSTTDSGHPNHLALTPSSGLRTRPSSIPKAWLRLWNKAFELLLGLPFSPCEGQVIEDEAVDNSGYSWL 0 | ||
2 | 0 2 | ||
0 | 1 YVNGTQDEKEQNLVFFQYHRQIFYQTCYAVWPGCQLLVWYRDECGQELGIKWDNRGKKEFe 1 | ||
1 | 2 EPKPEAHPCPSCPLAFSSEKFLSQHMKHNHPSQSSPETPERKHLQPEDPHPGHQNQQQQQHSDPHRWNDKAEGQQTGDRSKPMFENIRQEVTSRAFSSLPKGQMVCSREGNRMMETEPSPGLKVNPEVTGKLFLGVESSRIAK | ||
2 | VKYRGCGRDFSDRSHQSGHQRRHQ | ||
KKP | |||
SVCKKVKREFSHKSVLITHQRTHSGEKS | |||
YVCKESGRGFSAKSNLIRPRRTHTGEKP | |||
YVCGERGG.FSVSGLII.HQRAHSPEKP | |||
YVCREGRRGFGDKSSFIKHQRATLGEKS | |||
YVCKESGRGFS AKSNLIRPRRKKCRHDTTPHPQL | |||
>PRDM9_triMan Trichechus manatus (manatee) AHIN01064530 synteny: none (12740 bp) contig terminates prior to repeat | |||
0 MSPARATEESPGGDARRTPT 0 | |||
0 AKDAFRDISIYFSKEEWAEMGEWEKFRYRNVKRNYEALVAI 1 | |||
2 GLRAPRPAFMCHRRQAIKAQVDDTEDSDEEWTPRQQ 1 | |||
2 VKPPWVVSRVEQSKHQK 0 | |||
0 GTPRAPLNNESSLKEVSGTEILLSTAGSEQAQKLVSSPGEASTSDQHSRQKL 1 | |||
2 EPRRKEVEVKMYSLRERKGLAYQEVAEPQDDDYL 1 | |||
2 yCEKCQNFFIDACAVHGAPTFVKDSPADRGHPNRSALTLPPGLGIGPSGIPKAGLGVWNEASELPLGVHFGPYEGQVTEDEEAANSGYSWL 0 | |||
0 ITKGRNCYEYVDGKDESWANWMR 2 | |||
1 YVNCARNEEEQNLVAFQYHRQIFYRTCSTIQPGCELLVWYGDEYGQELGIKWGSRWKKELTSGR 1 | |||
2 EPKPEIHPCPSCPLAFSSQKFLSQHVKHRHPSQPFSGTPARKHLQPEDPRPGDQRQQHSERTQNDKAEDRETGDGSKRVFERTREGETSKVDSSLPKGQIGSSREGNRMMETEPSPGQKVNPEDTEKLLLGVGISRIVK | |||
VRHGECGQGFSQKSVLITHQRTHSGEKP | |||
YVCRECGRGFS | |||
> | >PRDM7_triMan Trichechus manatus (manatee) pseudogene AHIN01061278 internal stop and frameshift | ||
0 | 0 0 | ||
2 | 0 AKDAFRDISIYFSKEGWEEMGEWEKFRYRNMKRN^VQRNYKALVTI 1 | ||
2 | 2 GLKVPHPAFMCH*RQSIKSQTDDTEDSHEEWASRQQ 1 | ||
0 | 2 0 | ||
0 GILRASLSNKSSLKELSGT-IMLSRAGPEQAQKSVLPPGEASTSDKHSRQKL 1 | |||
2 | 2 EPRRKEVEVKTYNL*ERKDLVYQEVS*PQDGDYL 1 | ||
2 yCEKCQNF-TDSCAAHGDPTFVKDSAMDSGHPHHS------GLGIGPSSIPKARLEVWNKA GLHFSPYEGQVTEEEEAANSSYSWV 0 | |||
0 2 | |||
1 YVNYTQDKE*QNLVAFQYHRQIFYRTCRAIWPGCELLVWYGDEYGQELDIKWNSR-QKEFTAGR 1 | |||
> | >PRDM7_echTel Echinops telfairi (tenrec) genome Afro pseu 5 noDet 2 frameshifts plus stop codon | ||
0 | 0 0 | ||
2 | 0 1 | ||
2 | 2 GLRAPRPAFMCHHRPAAKGQVEDSEDSDEEWTPRQR 1 | ||
0 | 2 0 | ||
1 | 0 GMPGVSLRNESNLKVLSGTAILLTAAEPEQPH PGSPPGEATTSHEHLRQKV 1 | ||
2 | 2 epELRRRAVMMNSLRERKNLMYQEVSTPCDDNCL 1 | ||
2 YGERCHNFFIDTHIAHGATTFVKDS PMDRSNCSILPPGLRIGPSGIPEAGLGVWNEASELPLGLHFVPYEGQVTKDEAATNSGYSWM 0 | |||
0 ITKGRNCYEYVDGKDKSWANwMr 2 | |||
1 1 | |||
2 EPKPEVNPCPSCPLALSSQQLKHSHPFQSLPGTPAEKHLQAEDFHPRGQKLHHFEHHIRNERAEGLETGDGSKPMLERTRLGKMSKTTYNSPKGQTRSSGETNRIREADLNPGQGVNAEDTRNLFLGIGISRIAK | |||
VRCRECGHGFSVKSSLITHQRIHTGEKP | |||
YVCSECGQGFSQKSVLIRHQRIHTGEKP | |||
YICRECDRGFSRKSHLIKHQRTHSGEKP | |||
YVCRECGQGFSQKSVLITHHRTHSGEKP | |||
YVCRECGRGFSQKSDLIKHERTHS | |||
> | >PRDM7a_proCap Procavia capensis (hyrax) ABRQ01227339 Afro pseu 17 noDet frameshift and two stop codons in exon 10 | ||
0 | 0 0 | ||
2 | 0 AKDAFRDISIYFSKEEWAEMGEWEKSRYRNVKRNYEALVAI 1 | ||
2 | 2 GLRAPRPAFMCHRRQAIKAQVDNTEDSDEEWTPRQQ 1 | ||
0 | 2 AKPRSVASREELRKPQK 0 | ||
1 | 0 GTPKALLGNESSLKEVSGTAILLNTTGSEQAQKPVSSPGEASTSDQPSRWKL 1 | ||
2 | 2 EPRRKEAEVKRYNLREGTNPAYQEVGDTQDDDYL 1 | ||
2 yCEKYQKFCTDVCPAHGALAFLKDLSVERGHPKHSALTLPPGLRIGASGIPEAGLGVWSEASELPPGLHFGPCERQVTKDNEAANRGYLWP 0 | |||
0 ITKGRSCSLYMDRKDESRANWMR 2 | |||
1 YVRHAGDKEEQNLVAFQYHRQIFYRTCRPVQPGCELLVWPGAEDGQELGLQRGSRWKKELASQT 1 | |||
2 EARPEIHPCPSCPLAFSTPKFLSHHVKHSHPCQPFPGTLARRPLQPEDPHPGDRRQQHSEQPNWNDKAEGPEIGHVSRPVFEKTRQEGFSEARSSLPKGQMGRSREAERTTETQNSPGQKVNPEDTEILFLRGGISEIAK | |||
VKCGECGQGFSRKSHLIRHQRTHSGMKP | |||
YVCRECRRGFGVKSLLTRHQRTCSGMKP | |||
YVCRECGQGFRWKSHLIRHQRTHSGEKP | |||
FVCSECGRGFSVRSHLFTHQRTHSGEKP | |||
YVCKECGRGFSVKSYLTTHQRTHTGEKP | |||
YVCKECGRGFSWKSHLITHQRTHSGEKP | |||
YVCRQCGRGFSVQSHLIIHQRTHSGDKP | |||
YICRECGRDFTEKSSLIRHRRTHSGEKP | |||
YVCRDCG<font color=red>*</font>GFTRKSLLITHQRTHSGEKP | |||
YVYRECGRGFSCKSYLISHQKTHLGEKP | |||
YVCSDCGRGFSVKSQLVSHKRTHSGEKP | |||
FVCREC<font color=red>*</font>RGFSVKSSLISHQRTHSGEKP | |||
FVCRECGRGFSVKSSLIKHQRTHSGEKP | |||
YVCKECGRGFSQKSSLITHQRTHSGEKP | |||
YVCRECGRGFGLKSYLITHQRTHTGEKP | |||
YICRECG<font color=red>*</font>GFSVKSSLITDQRTHTGEKP | |||
YVCRECGRAFSKKSSLISHHRTHPAEAV | |||
YVHRECG | |||
> | >PRDM7b_proCap Procavia capensis (hyrax) ABRQ01392668 Afro pseu 13 noDet CpG stop in ZNF1, 4aa insert exon 4, frameshift exon 5 c to cc, 4aa del exon 9 etc | ||
0 | 0 0 | ||
1 | 0 AKEYFRDISMFFS<font color=red>*</font>ERWVEMSESEKFCYRNMKRNCETTGAG 1 | ||
2 | 2 GIRVFHPAFMIHPRKTIKAQMDDSEDSDEDWTARQQ 1 | ||
0 | 2 AKPPSVASREELRKPQK 0 | ||
1 | 0 GPSRAPLRIKSSLKRVSEPAIVWSTADSEQAQERVQKPVLSRREASASDQPLRRKV 1 | ||
2 EPRRHEAEDKRYSLRGGTGPACQEVGEPQDDDYL 1 | |||
2 yCEECRNFFIDTCVAHGTPVFIKDISVERGHPNRLALTLPTGLRIGPSSIPDAGLGVWNEASELPPGLHFGPCEGQVTEDEEAANSGYSWL 0 | |||
0 VTKGRSCFEYVDGKNEALANWMR 2 | |||
1 YVRRARDTEERNLVAFQYHRQIFYRTCCTVRPGCELLVWRGAEDSQALGSRRTMELTSQK 1 | |||
2 EARPEIHPCPSCPLAFSTQKFLSYHVNHSHSSEPFPGTHARRHLPREDPRPGYERDQRSEQHNWNDSTGGPERDVSRP VIERTWEGEISEACSSLPRGHMGRSREGERMAETQSSPGLKVTLAK | |||
VRWDEYGQGFGPKSHHITQQTKHSGKKP | |||
CVCKECG*GFRVKSLLKSHQMTHSGEKP | |||
YVCRECGRGFSVKSTLITHQRTHSGEKP | |||
YVCRECGRGFSVKSFLISHQRTHSGEKP | |||
YVCRECGRGFSWKSGLITHQRTHTGEKR | |||
YVCRECGHGFNRPSRLIRHQRTHSGEQP | |||
YVCRECGHGFNRRSQLIRHQRTHTGEQP | |||
YVCRECGQGFSGKSGLNRHQRTHSGEKP | |||
YVYKECGRGFSVKSTLIKHQRGHSGEKP | |||
YVCKECGRGFSRNSGLITHQRTHSGEQP | |||
YVCRECGRGFNQKSGVISHQRIHSGEKP | |||
FVCGECGRRFSWQSNLITHQRTHSGEKP | |||
FVCRECGRGFSAKTSLINHQRIH<font color=red>*</font>GKKP YVCRDGG | |||
> | >PRDM7_dasNov Dasypus novemcinctus (armadillo) AAGV020462211 9 xena pseu TRAPP | ||
0 | 0 0 | ||
0 AQDAFRDISTYFSREEWAEMGRWEKLRYRNVKRNYEALLAI 1 | |||
2 | 2 GLRAPRPAFMCHRKQSIKPQVDDAEDSDEEWTPRQQ 1 | ||
2 | 2 0 | ||
0 1 | |||
2 | 2 EPRRKGIDVKMYSLRERKGLAYEEVSEPQDDDYL 1 | ||
2 | 2 yCEKCQNFFIDSCTVHGPPIFVKDSAVDKGHPNRSALTLPSGLRIGPSGIPEAGLGIWNEASDLPLGLHFGPYEGQVTEDEEAANSGYSWL 0 | ||
0 | 0 ITKGRNYYEYEDGKDKSWANWMR 2 | ||
1 | 1 YVNCAWDDKEQNLVAFQYHRQIFYRTCRTIRPGCELLVWYGDEYGQELGIKWGSKWKKEFMTGT 1 | ||
2 | 2 ELKPEIHPCPSCPLAFSSEKFLSQHVRRHHPSQSFPAACAREHFQPQNPRPRGEEQQQHSDQCGWKDKAEGQETENRPKPLFERIKPMGSPRAFYNPPRGQMRSSREGKRMMEIQPSQDQKMNSE RGQLFLGVGIFKTEV | ||
IKFGENRQDFSDKSDHTSHQRTHTGEKP | |||
YVCRECGRGFSNNSHLTRHQRTHTGVKP | |||
YVCRECGQGFSVKPALTKHQRTHTVEKP | |||
yVCSECG GFSVKSTLITHQRTHTGEKP | |||
CVCRECGRGFNNKPDLTKHQRTHTGEKS | |||
YVCRECG GFSVKSTLIIHQRTHTGEKP | |||
YVCRECGRGFSEKSNLTVHQRTHTGEKP | |||
YVCRECGRGFSEKSNLTVHQRTHTGEKP | |||
YVCRECGRSFSVKSTLITHQRTHTVEKP | |||
YVCMKSEVVVSNKSHLNSHRRMKCGHRT PPPPQL | |||
> | >PRDM7_choHof Choloepus hoffmanni (sloth) ABVD01893961 2 xena gene noDet 0 0 | ||
0 | 0 1 | ||
2 | 2 1 | ||
0 | 2 0 | ||
0 | 0 1 | ||
2 | 2 1 | ||
2 ycekcQNFFFENCAAHGPPTLLKDSAVGQGRPKHSALVLPPGLRLGPSGIPEAGLGVWNEASDLPLGLHFGPYEGQVTEDEEATNSGYSWL 0 | |||
0 ITKGRNCYEYVDGKDKSCANWMR 2 | |||
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRAIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAEK 1 | |||
2 GLKPEIHPCPSCPLAFSTEKFLSQHVQRNHPSQIFPVTYARKHLQPQDPRPGDQQQPQPHSDQCHCSDKAEDQETEKRSKPLFESTKQMGISRAYSSPPEGQMRSSREDKRTMEIEPSQDQKMNPEETRLFVGVGILKTAR | |||
IKCGEYGQGFSVKPNLTTHQRTHTEEKP | |||
YVCRECGRGFGQKPNLSRHQRTHTGEKP | |||
YVCRECGRGFG | |||
> | >PRDMx_monDom Monodelphis domestica (opossum) gene genome no GAS8 fragment KRAB SSXRD SET weak C2H2 domain | ||
0 | 0 0 | ||
0 GEDAFKDISTYFSKKQWVKLKEWEKVRLKNVKRNYEAMIKI 1 | |||
2 | 2 GLSVPRPAFMCRGRQNKKVKVEESGDSDEEWIPKQL 1 | ||
0 | 2 VKTLRFPSRAKQRTHPK 0 | ||
0 1 | |||
2 | 2 DCRRKDVEVHIYSLRERKYQVYQEMWDPQDDDYL 1 | ||
1 | 2 yCEECQIFFLDSCPLHGPPTFVQDSAMVKGHPYCSAITLPPGLRIGLSGIPGAGLGVWNEASTLPLGLHFGPYKGKMTEDDEAANSGYSWM 0 | ||
0 ITKGRNCYEYVDGKEESCSNWMR 2 | |||
1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKRPLPELTGE 1 | |||
> | 2 GKPGISLCPSTLWASPLIPSSINTRCSKQPP<font color=red>*</font>VFLDSGTGKL<font color=red>*</font>AGRSTAGPATSNRFQLLSDKETSPKEHPSSLWGKTKQVDRREKFSLPQSQQVRGKESSSGEDLSRIQGKSTRQTTMAFQERNR | ||
0 | KECE<font color=red>*</font>GFTHQTNLVTHRWTHSGERP | ||
2 | YVCV<font color=red>*</font>GFTQKLGFSPYTWTL | ||
2 | |||
>PRDMx_macEug Macropus eugenii (wallaby) ABQO010244377 ABQO010410412 ABQO011136158 ABQO010410657 | |||
0 0 | |||
0 GEDTYKDISMYFSKKQWMELREWEKIRLKNVKQNYEAMIKI 1 | |||
2 gFSAPRPTFMCHGKQNKEAKVEESGDFDEEWIRKQP 1 | |||
2 0 | |||
0 1 | |||
2 ECRRKEAEVHIYNLRERKYQVYQEIWDPQDDDYL 1 | |||
2 FCEECQTFFLETCAVHGPPKFVQDSVMVKGHPYCSAITLPPGLRIGLSGIPGAGLGIWNEASNLPLGLHFGPYEGQMTEDDEAANSGYSWM 0 | |||
0 2 | |||
1 YVNCARDEEEQNLVAFQYHRKIFYRTCQIIRPGCELLVWYGDEYGQELGIKWGSKWKRPPITLT 1 | |||
2 espGIHVCPFCPLGSPLMHSQSTYAAQTSPQICLDSRTRNNYEPDQLLPPSSSCVSDKVEISQKQRPSSLCGKTKQVNLVEMLSLPQSPQVSKKSSSMDWDVSRIQGKSAKQTTQGFQKGDKKGFGS | |||
YKCGEYKQGFTSKSVLNRHRQKHSGKKP | |||
YVCEECGRGFTQVSNLTTHRQTHSGEKP | |||
YVCEECGRGFARKLNLTTHRRTHSGEKP | |||
YVCEECGRGFTQGSSLITHRRTHSGEKP | |||
YVCEECGRGFAWKLNLTTHRRTHSGEKP | |||
YVCKECGRGFTQGSSLITHRRTHSGEKP | |||
YVCKECGRGFTQGSNLTTHRRTHSGEKP | |||
YVCKECGRGFAWKSNLTTHRRTHSGEKP | |||
YVCKECGRGFTQVSNLIAHRRTHSGEKF | |||
YVYGQE FTWKSDLSTCR | |||
> | >PRDMx_sarHar Sarcophilus harrisii (tasmanian_devil) AFEY01386448 distal frameshifts and stop codons, syntenic -PSMC4 | ||
0 | 0 0 | ||
2 | 0 EEDSFKDISMYFSKKQWMELRDWEKVRFKNVKRNYEAMIKI 1 | ||
2 | 2 GLTASRPTFMCRGKQNRRAKVEESGDSDEEWMPKQL 1 | ||
2 VKASRFSSRLKQKTHLR 0 | |||
0 1 | |||
2 eCRKKDAAVHIYNLRERKYPIYQEIWDPQDDDYL 1 | |||
2 FCEECQTFFLETCAVHGPPKFVQDGAMIKGYPYCSAITLPPGLRIGLSGIPNAGLGVWNEGSNLPMGLHFGPYEGKSTEDDEAANSGYSWM 0 | |||
0 ITKGRNSYEYVDGKEESCSNWMR 2 | |||
1 YVNCAREEEEQNLVAFQYQRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGRKWKRPLTGIT 1 | |||
2 GIHLCPSCPSDFSTHAFLSQQVPKQPSQGFLDSTTGSHGLGNLHPDQLLPPGYSCVSDKAETSRKEHPSTLWEKIKKVDLEEPASLPQRQHVREEESNLGEWDLSRIQGESVKLTSLALQEESQEGLGQ | |||
YKCGEGKQRYSSKPGLIRHRQRTHSGEKC | |||
YVCEECKRGFARRSYLNIHRRRHSGQKP | |||
HVCEECKRGFADKSTLIRHRWTHSKEKP | |||
YICEECKQGFTQKSYLIKHRWKHLGEKP | |||
YVCKECKQRFTQRSYLNTHRWRHRQRSL | |||
<font color=magenta>v</font>FC<font color=magenta>a</font>ECR<font color=red>*</font>GFT<font color=red>*</font>RS<font color=red>*</font>LIIHRWTHSGERP | |||
> | YVCEECKGGFTQRSYLN<font color=magenta>t</font>HTDGNVGKEEP | ||
0 | YVCEECR | ||
2 | |||
0 | >PRDMxa_ornAna Ornithorhynchus anatinus (platypus) gene genome chrX5 fragment X5 +- 20577549 no iMet possible in first exon phase 2 | ||
2 | 0 0 | ||
0 1 | |||
2 1 | |||
2 0 | |||
0 1 | |||
2 RIGKKPQVRDFNLRKQKRKIYNENYRPEDDDYL 1 | |||
2 yCEICQTFFLEKCVLHGPPVFVQDLPVEKWRPNRSTITLPPGMQIKVSGIPNAGLGVWNQATSLPRGLHFGPYMGIRTKNEKESHSGYSWM 0 | |||
2 IVRGKNYEYLDGKDKAFSNWMR 2 | |||
1 YVNCARSEREQNLVAIQYQGEIYYRTCRVIPPGQELLVWYGLEYGRHLGILPNNNNPEP 1 | |||
2 ERAKARVRKSERIEKAMARVRKSEQIERAKARVRTSERIERAMATV RKSERIERAKVTVKKSEQIERAMGRVRKSERIERAKDMGRKKALGGLPRPCRGGLSDETQQRKGGGHEQLGQKPGPSEA RAGPAEGSATPRR | |||
HCCDVCRKAFKRLSHLRQHKRIHTGEKP | |||
LVCKVCRRTFSDPSNLNRHSRIHTGLRP | |||
YVCKLCRKAFADPSNLKRHVFSHTGHKP | |||
FVCEKCGKGFNRCDNLKDHSAKHSEDNSTPKP | |||
> | >PRDMxb_ornAna Ornithorhynchus anatinus (platypus) gene genome chrX5 tandem fragment slight frameshift taa to ta YVN exon X5 +- 20605294 20611704 no iMet possible in first exon phase 2 gg as expected | ||
0 | 0 0 | ||
0 1 | |||
0 | 2 1 | ||
2 | 2 0 | ||
0 1 | |||
2 RSGKKPQVRDFNLRKQKRKMYTEESEPEDDDYL 1 | |||
2 yCEDCQTFFLEKCSVHGPPVFVQDCEAKRCQQNRSEVTLPPGLLIKMSGIPNAGLGVWNQATSLPRGLYFGPFVGIRKNNVKDSLSGYSWA 0 | |||
0 ILRGRNYEYLDGKNTSFSNWMR 2 | |||
0 | 1 YVNCPRTKYEQNLVAIQYHREIYYRTTPCDSTRSRVAGVVWRRVRSYLGIFWKSETPKS 1 | ||
2 | 2 ERPHSSGGSFAPSARSGGVKQRIWSKRRSAALQRTRERRNSTHDFPPKHEDTAARQDERQCPDRGRAKQRGVRKSEQIERAKAMGRKKALGGLSPPRRERLSDEAGQRKKSGHEQFWQKPGPSEAWAGPAEGSTIPRR | ||
HCCDVCGKAFNRLSRLKQHKRVHTGEKP | |||
LVCKICKRAFSDPSNLNRHAKRHTGEKP | |||
FVCRVCGRSFNRSDNMNEHRWKHTSNNIIP NTGHMSATVVENASLCINRNYQIYKERATYL | |||
2 | |||
> | >PRDMx_danRer Danio rerio (zebrafish) Q6P2A1 transcript BC064665 no KRAB SSXRD or exon 5 but knuckle SET early ZNf C2H2 array | ||
0 MSLSP 1 | |||
2 DLPPSEEQNLEIQGSATNCYSVVIIEEQDDTFNDQPF 1 | |||
2 YCEMCQQHFIDQCETHGPPSFTCDSPAALGTPQRALLTLPQGLVIGRSSISHAGLGVFNQGQTVPLGMHFGPFDGEEISEEKALDSANSWV 0 | |||
0 ICRGNNQYSYIDAEKDTHSNWMK 2 | |||
1 FVVCSRSETEQNLVAFQQNGRILFRCCRPISPGQEFRVWYAEEYAQGLGAIWDKIWDNKCISQ 1 | |||
2 GSTEEQATQNCPCPFCHYSFPTLVYLHAHVKRTHPNEYAQFTQTHPLESEAHTPITEVEQCLVASDEALSTQTQPVTESPQEQISTQNGQPIHQTENSDEPDASDIYTAAGEISDEI | |||
HACVDCGRSFLRSCHLKRHQRTIHSKEKP | |||
YCCSQCKKCFSQATGLKRHQHTHQEQEKNIESPDRPSDI | |||
YPCTKCTLSFVAKINLHQHLKRHHHGEYLRLVESGSLTAETEEDHT | |||
EVCFDKQDPNYEPPSRGRKSTKNSLKGRGCPKKVAVGRPRGRPPKNKNLEVEVQKIS | |||
PICTNCEQSFSDLETLKTHQCPRRDDEGDNVEHPQEASQ | |||
YICGECIRAFSNLDLLKAHECIQQGEGS | |||
YCCPHCDLYFNRMCNLRRHERTIHSKEKP | |||
YCCTVCLKSFTQSSGLKRHQQSHLRRKSHRQSSALFTAAI | |||
FPCAYCPFSFTDERYLYKHIRRHHPEMSLKYLSFQEGGVLSVEKP | |||
HSCSQCCKSFSTIKGFKNHSCFKQGEKV | |||
YLCPDCGKAFSWFNSLKQHQRIHTGEKP | |||
YTCSQCGKSFVHSGQLNVHLRTHTGEKP | |||
FLCSQCGESFRQSGDLRRHEQKHSGVRP | |||
CQCPDCGKSFSRPQSLKAHQQLHVGTKL | |||
FPCTQCGKSFTRRYHLTRHHQKMHS | |||
>PRDMx_salSal Salmo salar NM_001173912 | |||
0 MESEWKSGGEEESGSEGERTPSSSHRDP 1 | |||
2 VCVSEQMKRAWLRQMNLRSRARVGYTEEEELRDEEYF 1 | |||
2 FCEECKSFFIEECELHGPPLFIPDTPAPLGAPDRARLTLPPGLEVRTSAIPGAGLGVFNHGHSVTQGTHYGPYEGELTDKELDMESGYSWV 0 | |||
0 IYKSKQRDEYIDGKRDTHSNWMR 2 | |||
1 YVNCARSEDEQNLVAFQYRGGILYRCCKPIAVGEELLVWYGEKYARDLGIVFDFLWDKKCSAR 1 | |||
2 GVNESSQSQIFSCSGCLFSFTAQTYLYKHIKRCHREECVRLPRSGGIRAETLAPPSGSQRCSTTPDRTPITLLTQKHRDTGKPAP | |||
HHCSQCGKSFRRSGDLKVHQRTHTGERP | |||
YHCSQCGKRFSVSGHLKTHQRTHTGERP | |||
YHCSQCGKSFCRSGDLKVHQRTHTGERP | |||
YHCSQCGKRFSVSRHLKRHQHIHTGERP | |||
YHCSQCGKSFSASWSVKRHQITVHSVGRVSVSQEA | |||
>PRDMx_oncMyk Oncorhynchus mykiss testis FP324541 CR372724 | |||
0 mTPSSSHRDPVC 1 | |||
2 VSEQRKRAWLKQVNLCSRARVRVGYTEEEELREEDYF 1 | |||
2 FCEECKSFFIEECELHGPPLFIQDTPAPLGAPDRARLTLPPGLEVRTSAIPGAGLGVFNYGHSVTQGTHYGPYEGELTDTELAMESGYSWV 0 | |||
0 IYKSKQSDEYIDAKRETHSNWMR 2 | |||
1 YVNCARNEEEQNLVAFQYRGGILYRCCKPLAVGEELLVWYGEEYARDLGIIFDFLWDRKSSAR 1 | |||
2 GVNESSQSQIFSCSGCPFSFTAQIYLYKHTKRCHREEYVRLPRSGGIRSETLAPPSGSQRCSTTPDRTPITLLTQKHQDTGKPRP | |||
HHCSQCGKSFHRSGDLKVHQRTHTGERP | |||
YHCSQCGKRFSVSGNLKTHQRIHTGERP | |||
YPCSQCGKSFHRSDLKVHQRTHTGEKP | |||
YHCSQCGKRFSVSGNLKTHQRIHTGERL | |||
YPCSQCGKSFHRSELKVQQRTRPGKKTISLFPVWE | |||
>PRDMx_ictPun Ictalurus punctatus FD367165 FD063496 C-terminus missing second gene present | |||
0 MKTEAKDGGTEGI 2 | |||
1 VKKETLELSISNHGNSFHIIPEVVSIKEEEADVKDFL 1 | |||
2 YCEVCKSVFFSKCEVHGPALFIADSPVPMGVADRARQTLPPGLEIQKSGIPDAGLGVFNKGETVPVGAHFGPYQGELVDKEEAMNSVYSWV 0 | |||
0 IYMSRQCEKYIDAKREVHANWMR 2 | |||
1 YVNCAHSDGEQNLVAFQYRGGILYRCCRPINPGQELLVWYEEKYASDVGPIFAQLWNIKCSLSGKVHT | |||
=== Other genes of relevance === | |||
It is instructive to consider certain closely related placental KRAB, ZNF and PRDM genes that may have some connection to the origin of PRDM7 and PRDM9. Nomenclature is very unsatisfactory in these gene families, as can be seen from lack of correspondence between gene name and intronation which is exceedingly well conserved in metazoa. For example, HKR1 a conventional ZNF family member, is egregiously misnamed. The methylase component is exceedingly old with clear antecedents in bacteria. Evidently gene duplications in an early intronless stem eukaryote were subsequently intronated randomly in different paralogs and shuffled into various larger proteins. Within PRDM*, the gene tree is (((PRDM7/9,PRDM11),(PRDM4,PRDM10)),PRDM6) with others only related by a PR (SET) domain. PRDM11 and its novel giant terminal exon are discussed in a [[PRDM11:_giant_missing_exon|separate article]] | |||
A set of fragmentary sequences from murid rodents is also of some comparative interest. These include common strain variants of lab mouse as well as close relatives. Only the terminal zinc finger array is available for most of these. While these are likely PRDM7 (rather than PRDM9 which rodents never had), it is not possible to decisively establish this with GAS8 synteny in any of the rodents (or lagomorphs) currently the subject of a genome project. | |||
>PRDM11_homSap Homo sapiens (human) corrected 511+722 aa <font color=#00CC66>PhosS</font> <font color=blue>3RAY coverage</font> <font color=#FFBB66>knuckle</font> <font color=red>SET</font> <font color=#990099>ZnF_TTF</font> <font color=brown>hATC_dimerization</font> no early zinc finger or terminal array syn PFM8 related ZNF 862 | |||
0 MTENMKECLAQTNAAVGDMVTVVKTEVC<font color=#00CC66>S</font>PLRDQEYGQPC 2 | |||
1 SRRPD<font color=blue>SSAMEVEPKKLKGKRDLIVPKSFQQVDF</font><font color=#FFBB66>W 1 | |||
2 FCESCQEYFVDECPNH</font><font color=blue>GPPVFVSDTPVPVGIPDRAALTIP</font><font color=red>QGMEVVKDTSGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 | |||
0 IVDKNNRYKSIDGSDETKANWMR 2 | |||
1 YVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKR</font><font color=blue>LHSMSQETIHRNLAR 1 | |||
2 GEKRLQREKSEQVLDNPEDLRGPIHLSVLRQGK</font>SPYKRGFDEGDVHPQAKKKKIDLIFKDVLEASLESAKVEAHQLALSTSLVIRKVPKYQDDAYSQCATTMTHGVQNIGQTQGEGDWKVPQGVSKEPGQLEDEEEEPSSFKADSPAEASLASDPHELPTTSFCPNCIRLKKKVRELQAELDMLKSGKLPEPPVLPPQVLELPEFSDPA 1 | |||
2 ASESMVSGPAIMEDDDQEVDSADESVSNDMMTATDEPSKMSSATG<font color=#990099>RRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM</font> | |||
CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSVILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAF | |||
SALGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTAATLCEETEFLGDIRAVRWIIGEQN | |||
VLNALIKDYLEVVAHLKEVSSQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYIFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAMKNLRVAE | |||
AKFQSIREKICQKTQVILAQRFDSRSRIFVKACQVFDLAAWPRSSEELMSYGKEDMVQIFDHLEAIPT<font color=brown>FSRDVCREGLDPRGSLLMEWRELKADYYTKNGFKDLISHICKYKQRFPLLNK | |||
IIQVLKVLPTSTACCEKGRNALQRVRKNHRSRLTLEQLSDLLTIAVN</font>GPPITNFDAKRALDSWFEEKSGNSYALSAEVLSRMSALEQKPALQTMDHGTEFYPDI* 0 | |||
>PRDM4_homSap Homo sapiens (human) 801 aa knuckle, SET, early zinc and array | |||
0 MHHR 2 | |||
1 MNEMNLSPVGMEQLTSSSVSNALPVSGSHLGLAASPTHSAIPAP 1 | |||
2 GLPVAIPNLGPSLSSLPSALSLMLPMGIGDRGVMCGLPERNYTLPPPPYPHLESSYFRTILP 1 | |||
2 GILSYLADRPPPQYIHPNSINVDGNTALSITNNPSALDPYQSNGNVGLEPGIVSIDSRSVNTHGAQSLHPSDGHEVALDTAITMENVSRVTSPISTDGMAEELTMDGVAGEHSQIPNGSRSHEPLSVDSVSN | |||
NLAADAVGHGGVIPMHGNGLELPVVMETDHIASRVNGMSDSALSDSIHTVAMSTNSVSVALSTSHNLASLESVSLHEVGLSLEPVAVSSITQEVAMGTGHVDVSSDSLSFVSPSLQMEDSNSNKENMATLFT<font color = red>I</font> 1 | |||
2 <font color = red>WCTLCDRAYPSDCPEH</font>GPVTFVPDTPIESRARLSLP<font color = purple>KQLVLRQSIVGAEV 1 | |||
2 GVWTGETIPVRTCFGPLIGQQSHSMEVAEWTDKAVNHIWK 0 | |||
0 IYHNGVLEFCIITTDENECNWMMFVRKAR 2 | |||
1 NREEQNLVAYPHDGKIFFCTSQDIPPENELLFYYSRDYAQQ</font>I 1 | |||
2 GVPEHPDV<font color = magenta>HLCNCGKECNSYTEFKAHLTSH</font>IHNHLPTQGHSGSHGPSHSKERKWKCSMCPQAFISPSKLHVHFMGHMGMKP | |||
<font color = blue>HKCDFCSKAFSDPSNLRTHLKIHT 12 GQKN</font> | |||
<font color = blue>YRCTLCDKSFTQKAHLESHMVIHTGEKN</font> | |||
<font color = blue>LKCDYCDKLFMRRQDLKQHVLIHTQ 21 ERQ</font> | |||
<font color = blue>IKCPKCDKLFLRTNHLKKHLNSHEGKRD</font> | |||
<font color = blue>YVCEKCTKAYLTKYHLTRHLKTCKGPTS</font> SSSAPEEEEEDDSEEEDLADSVGTEDCRINSAVYSADESLSAHK* 0 | |||
>PRDM10_homSap Homo sapiens (human) 1160 aa knuckle, SET, early zinc and array | |||
0 MDSKDESSHVWPTSAEHEQNAAQ 0 | |||
0 VHFVPDTGTVAQIVYTDDQVRPPQQVVYTADGASYTSVDGPEHTLVYIHPVEAAQ 0 | |||
0 TLFTDPGQVAYVQQDATAQQ 0 | |||
0 ASLPVHNQVLPSIESVDGSDPLATLQTPLGRLEAKEEEDEDEDEDTEEDEEEDGEDTDLDDWEPDPPRPFDPHD<font color = red>L</font> 1 | |||
2 <font color = red>WCEECNNAHASVCPKHGPLH</font>PIPNRPVLTRARASLPL<font color = purple>VLYIDRFLGGVFSKRRIPKRTQFGPVEGPLVRGSELKDCYIHLK 0 | |||
0 VSLDKGDRKERDLHEDLWFELSDETLCNWMMFVRPAQNHLEQNLVAYQYGHHVYYTTIKNVEPKQELK 0 | |||
0 VWYAASYAEF</font>VNQKIHDISEEERK 1 | |||
2 VLREQEKN<font color = magenta>WPCYECNRRFISSEQLQQHLNSH</font>DEKLDVFSR 2 | |||
1 TRGRGRGRGKRRFGPGRRPGRPPKFIRLEITSENGEKSDDGTQ 0 | |||
0 DLLHFPTKEQFDEAEPATLNGLDQPEQTTIPIPQLPQETQSSLEHEPETHTLHLQPQHEESVVPTQSTLTADDMRRAKRIR 0 | |||
0 LELQ 0 | |||
0 NAALQHLFIRKSFRP | |||
<font color = blue>FKCLQCGKAFREKDKLDQHLRFH</font>GREGNCP | |||
<font color = blue>LTCDLCNKGFISSTSLESHMKLH</font>SDQKT | |||
<font color = blue>YSCIFCPESFDRLDLLKDHVAIH</font>INDGY | |||
<font color = blue>FTCPTCKKRFPDFIQ</font> 00 <font color = blue>VKKHVRSFH</font>SEKI | |||
<font color = blue>YQCTECDKAFCRPDKLRLHMLRH</font>SDRKD | |||
<font color = blue>FLCSTCGKQFK</font> 00 <font color = blue>RKDKLREHMQRMH</font>NPEREAKKADRISRSKTFKPRITSTDYDSFT | |||
<font color = blue>FKCRLCMMGFRRRGML</font> 00 <font color = blue>VNHLSKRH</font>PDMKIEEVPELTLPIIKPNRD | |||
<font color = blue>YFCQYCDK</font> 00 <font color = blue>VYKSASKRKAHILKNH</font>PGAELPPSIRKLRPAGPGEPDPMLSTHTQLTGTIATPP | |||
<font color = blue>VCCPHCSKQYSSK</font> 00 <font color = blue>TKMVQHIRKKH</font>PEFAQLSNTIHTPLTTAVISATPAVLTTDSATGETVVTTDLLTQAMTELSQTLTTDYRTPQGDYQRIQYIPVSQSASGLQQPQHIQLQVVQVAS 0 | |||
0 ATSPHQSQQSTVDVGQLHDPQPYPQHAIQVQHIQVSEPTASAPSSAQ 0 | |||
0 VSGQPLSPSAQQAQQGLSPSHIQGSSSTQGQALQQQQQQQQNSSVQHTYLPSAWNSFRGY 1 | |||
2 SSEIQMMTLPPGQFVITDSGVATPVTTGQVKAVTS 0 | |||
0 GHYVLSESQSELEEKQTSALSGGVQVEPPAHSDSLDPQTNSQQQTTQYIITTTTNGNGSSEVHITKP* 0 | |||
>PRDM15_homSap Homo sapiens (human) 1507 aa knuckle, SET, early zinc finger and intronated array | |||
0 MPRRRPPASGAAQFPERIATRSPDPIPLCTFQRQ 0 | |||
0 PRAAPVQPPCRLFFVTFAGCGHRWRSESKPGWISRSRSGIALRAARPP 1 | |||
2 GSSPPRPAAPRPPPPGGVVAEAPGDVVIPRPRVQPMRVARGGPWTPNPAFREAESW 2 | |||
1 SQIGNQRVSEQLLETSLGNEVSDTEPLSPASAGLRRNPALPP 1 | |||
2 GPFAQNFSWGNQENLPPALGKIANGG 1 | |||
2 GTGAGKAECGYETESHLLEPHEIPLNVN 0 | |||
0 THKFSDCEFPYEFCTVCFSPFKLLGMSGVEGVWNQHSRSASMHTFLNHSATGIREAGCRKDMP 0 | |||
0 VSEMAEDGSEEIMF<font color = red>I</font> 12 <font color = red>WCEDCSQYHDSECPELGP</font>VVMVKDSFVLSRAR 2 | |||
1 SWPASGHVHTQAGQGMRGYEDRDRADPQQLPEAVPAGLVRRLSGQQLPCRSTLTWGRLCHLVAQGR 2 | |||
1 SSLP<font color = purple>PNLEIRRLEDGAEGVFAITQLVKRTQFGPFESRRVAKWEKESAFPLK 0 | |||
0 VFQKDGHPVCFDTSNEDDCNWMMLVRPAAEAEHQNLTAYQHGSDVYFTTSRDIPPGTELRVWYAAFYAKK</font>MDKPMLKQAGSGVH 1 | |||
2 AAGTPENSAPVESEPSQ<font color = magenta>WACKVCSATFLELQLLN</font> 12 <font color = magenta>EHLLGH</font>LEQAKSLPPGSQSEAAAPEKEQDTPRGEPPAVPESENVATKEQKKKPRRGRKPKVSKAEQPLVIVEDKEPT 12 EQVAEIITEVPPDEPVSATPDERIMELVLGKLATTTTDTSSVPK 21 FTHHQNNTITLKRSLILSSRHGIRRKLIKQLGEHKR | |||
<font color = blue>VYQCNICSKIFQNSSNLSRHVRSH</font> 12 GDKL | |||
<font color = blue>FKCEECAKLFSRKESLKQHVSYKH</font>SRNE 00 VDGEYR | |||
<font color = blue>YRCGTCEKTFRIESALEFHNC</font>RT 12 DDKT | |||
<font color = blue>FQCEMCFRFFSTNSNLSKHKKKH</font>GDKK | |||
<font color = blue>FACEVCSKMFYRKDVMLDHQRRH</font>LE 12 GVRRVKblueLEAGGENLVRYKKEP | |||
<font color = blue>SGCPVCGK</font> 00 <font color = blue>VFSCRSNMNKHLLTH</font>GDKK | |||
<font color = blue>YTCEICGRKFFRVDVLRDHIHVH</font>FK 00 DIALMDDHQREEFIGKIGISSEENDDNSDESADSEPHK | |||
<font color = blue>YSCKRCQ</font> 00 <font color = blue>LTFGRGKEYLKHIMEVH</font>KEKG | |||
<font color = blue>YGCSICNRRFALKATYHAHMVIH</font>RENLPDPNVQK 21 YI | |||
<font color = blue>HPCEICGRIFNSIGNLERHKLIH</font>T 12 GVKS | |||
<font color = blue>HACEQCGKSFARKDMLKEHMRVH</font>DNVRE | |||
<font color = blue>YLCAECGK</font> 12 <font color = blue>GMKTKHALRHHMKLH</font>KGIKE | |||
<font color = blue>YECKECHRRFAQKVNMLKHCKRH</font>T 12 GIKD | |||
<font color = blue>FMCELCGKTFSERNTMETHKLIH</font>T 12 VGKQ | |||
<font color = blue>WTCSVCDKKYVTEYMLQKHVQLTH</font>DKVEA | |||
<font color = blue>QSCQLCGTKVSTRASMSRHMRRKH</font>PE 0 | |||
0 VLAVRIDDLDHLPETTTIDASSIGIVQ 0 | |||
0 PELTLEQEDLAEGKHGKAAKRSHKRKQKPEEEAGAPVPEDATFSEYSEKETEFTGSVGDETNSAVQSIQQ 0 | |||
0 VVVTLGDPNVTTPSSSVGLTNITVTPITTAAATQFTNLQPVAVGHLTTPERQLQLDNSILTVTFDTVSGSAMLHNRQNDVQIHPQPEASNPQSVAHFINLTTLVNSITPLGSQLSDQHPLTWRAVPQTDVLPPSQPQAPPQQAAQPQVQAEQQQQQMYSY* 0 | |||
>PRDM6_homSap Homo sapiens (human) 595 aa knuckle, SET, no early zinc finger, short array | |||
0 MLKPGDPGGSAFLKVDPAYLQHWQQLFPHGGAGPLKGSGAAGLLSAPQPLQPPPPPPPPERAEPPPDSLRPRPASLSSASSTPASSSTSASSASSCAA | |||
AAAAAALAGLSALPVSQLPVFAPLAAAAVAAEPLPPKELCLGATSGPGPVKCGGGGGGGGEGRGAPRFRCSAEELDYYLYGQQRMEIIPLNQHTSDPN<font color = red>N</font> 1 | |||
2 <font color = red>RCDMCADNRNGECPMH</font>GPLHSLRRLVGTSSAAAAAPPPELPEWLRDLPRE<font color = purple>VCLCTSTVPGLAYGICAAQRIQQGTWIGPFQGVLLPPEKVQAGAVRNTQHLWE 0 | |||
0 IYDQDGTLQHFIDGGEPSKSSWMRYIRCARHCGEQNLTVVQYR 2 | |||
1 SNIFYRACIDIPRGTELLVWYNDSYTSF</font>FGIPLQCIAQDEN 1 | |||
2 LNVPSTVMEAMCRQDALQPFNKSSKLAPTTQQRSVVFPQTPCSRNFSLLDKSGPIESGFNQINVKNQRVLASPTSTSQLHSEFSDWHL | |||
<font color = blue>WKCGQCFKTFTQRILLQMHVCTQNPDR</font> 21 <font color = blue>P</font> | |||
<font color = blue>YQCGHCSQSFSQPSELRNHVVTHSSDRP</font> | |||
<font color = blue>FKCGYCGRAFAGATTLNNHIRTHTGEKP</font> | |||
<font color = blue>FK</font> 21 <font color = blue>CERCERSFTQATQLSRHQRMPNECKP</font> ITESPESIEVD* 0 | |||
>ZNF133_homSap Homo sapiens (human) 653 aa KRAB, early zinc finger and array | |||
0 <font color = green>MAFRDVAVDFTQDEWRLLSPAQRTLYREVMLENYSNLVSL 1 | |||
2 GISFSKPELITQLEQGKETWR</font>EEKKCSPATCP 1 | |||
2 DPEPEL<font color = magenta>YLDPFCPPGFSSQKFPMQHVLCNH</font>PPWIFTCLCAEGNIQPGDPGPGDQEKQQQASEGRPWSDQAEGPEGEGAMPLFGRTKKRTLGAFSRPPQRQPVSSRNGLRGVELEASPAQTGNPEETDKLLKRIEVLGFGT | |||
<font color = blue>VNCGECGLSFSKMTNLLSHQRIHSGEKP | |||
YVCGVCEKGFSLKKSLARHQKAHSGEKP | |||
IVCRECGRGFNRKSTLIIHERTHSGEKP | |||
YMCSECGRGFSQKSNLIIHQRTHSGEKP | |||
YVCRECGKGFSQKSAVVRHQRTHLEEKT | |||
IVCSDCGLGFSDRSNLISHQRTHSGEKP | |||
YACKECGRCFRQRTTLVNHQRTHSKEKP | |||
YVCGVCGHSFSQNSTLISHRRTHTGEKP | |||
YVCGVCGRGFSLKSHLNRHQNIHSGEKP | |||
IVCKDCGRGFSQQSNLIRHQRTHSGEKP | |||
MVCGECGRGFSQKSNLVAHQRTHSGERP | |||
YVCRECGRGFSHQAGLIRHKRKHSREKP | |||
YMCRQCGLGFGNKSALITHKRAHSEEKP | |||
CVCRECGQGFLQKSHLTLHQMTHTGEKP | |||
YVCKTCGRGFSLKSHLSRHRKTTS</font>VHHR LPVQPDPEPCAGQPSDSLYSL* 0 | |||
>HKR1_homSap Homo sapiens (human) 659aa KRAB, early zinc finger and array | |||
0 MRVNHTVSTMLPTCMVHRQTMSCSGAGGITAF<font color = green>VAFRDVAVYFTQEEWRLLSPAQRTLHREVMLETYNHLVSL 1 | |||
2 EIPSSKPKLIAQLERGEAPWR</font>EERKCPLDLCP 1 | |||
2 ESKPEIQLS<font color = magenta>PSCPLIFSSQQALSQHVWLSH</font>LSQLFSSLWAGNPLHLGKHYPEDQKQQQDPFCFSGKAEWIQEGEDSRLLFGRVSKNGTSKALSSPPEEQQPAQSKEDNTVVDIGSSPERRADLEETDKVLHGLEVSGFGE | |||
<font color = blue>IKYEEFGPGFIKESNLLSLQKTQTGETP | |||
YMYTEWGDSFGSMSVLIKNPRTHSGGKP | |||
YVCRECGRGFTWKSNLITHQRTHSGEKP | |||
YVCKDCGRGFTWKSNLFTHQRTHSGLKP | |||
YVCKECGQSFSLKSNLITHQRAHTGEKP | |||
YVCRECGRGFRQHSHLVRHKRTHSGEKP | |||
YICRECEQGFSQKSHLIRHLRTHTGEKP | |||
YVCTECGRHFSWKSNLKTHQRTHSGVKP | |||
YVCLECGQCFSLKSNLNKHQRSHTGEKP | |||
FVCTECGRGFTRKSTLSTHQRTHSGEKP | |||
FVCAECGRGFNDKSTLISHQRTHSGEKP | |||
FMCRECGRRFRQKPNLFRHKRAHSGA | |||
FVCRECGQGFCAKLTLIKHQRAHAGGKP | |||
HVCRECGQGFSRQSHLIRHQRTHSGEKP | |||
YICRKCGRGFSRKSNLIRHQRTHSG</font>* 0 | |||
>ZNF343_homSap Homo sapiens (human) 599aa KRAB, early zinc finger and array | |||
0 MMLPYPSALGDQYWEEILLPKNGENVETMKKLTQNHKAK 1 | |||
2 GLPSNDTDCPQKKEGKAQIV 0 | |||
0 VP<font color = green>VTFRDVTVIFTEAEWKRLSPEQRNLYKEVMLENYRNLLSL 1 | |||
2 AEPKPEI</font><font color = magenta>YTCSSCLLAFSCQQFLSQHVL</font>QIFLGLCAENHFHPGNSSPGHWKQQGQQYSHVSCWFENAEGQERGGGSKPWSARTEERETSRAFPSPLQRQSASPRKGNMVVETEPSSAQRPNPVQLDKGLKELETLRFGA | |||
<font color = blue>INCREYEPDHNLESNFITNPRTLLGKKP | |||
YICSDCGRSFKDRSTLIRHHRIHSMEKP | |||
YVCSECGRGFSQKSNLSRHQRTHSEEKP | |||
YLCRECGQSFRSKSILNRHQWTHSEEKP | |||
YVCSECGRGFSEKSSFIRHQRTHSGEKP | |||
YVCLECGRSFCDKSTLRKHQRIHSGEKP | |||
YVCRECGRGFSQNSDLIKHQRTHLDEKP | |||
YVCRECGRGFCDKSTLIIHERTHSGEKP | |||
YVCGECGRGFSRKSLLLVHQRTHSGEKH | |||
YVCRECRRGFSQKSNLIRHQRTHSNEKP | |||
YICRECGRGFCDKSTLIVHERTHSGEKP | |||
YVCSECGRGFSRKSLLLVHQRTHSGEKH | |||
YVCRECGRGFSHKSNLIRHQRTH</font>* 0 | |||
>ZNF589_homSap Homo sapiens (human) 364 aa KRAB, early zinc finger and truncated array | |||
0 MWAPREQLLGWTAE 1 | |||
2 ALPAKDSAWPWEEKPRYL 0 | |||
0 GP<font color = green>VTFEDVAVLFTEAEWKRLSLEQRNLYKEVMLENLRNLVSL 1 | |||
2 AESKPEV</font><font color = magenta>HTCPSCPLAFGSQQFLSQDELHNH</font>PIPGFHAGNQLHPGNPCPEDQPQSQHPSDKNHRGAEAEDQRVEGGVRPLFWSTNERGALVGFSSLFQRPPISSWGGNRILEIQLSPAQNASSEEVDRISKRAETPGFGAVTFGECALAFNQKSNLFRQKAVTAEKSSDKRQS | |||
<font color = blue>QVCRECGRGFSRKSQLIIHQRTHTGEKP | |||
YVCGECGRGFIVESVLRNHLSTHSGEKP | |||
YVCSHCGRGFSCKPYLIRHQRTHTREKS | |||
FMCTVCGRGFREKSELIKHQRIHTGDKP | |||
YVCRD</font>* 0 | |||
>ZNF169_homSap Homo sapiens (human) 603aa KRAB, no early zinc finger but array | |||
0 MSPGLLTTRKEAL<font color = green>MAFRDVAVAFTQKEWKLLSSAQRTLYREVMLENYSHLVSL 1 | |||
2 GIAFSKPKLIEQLEQGDEPWR</font>EENEHLLDLCP 1 | |||
2 EPRTEFQPSFPHLVAFSSSQLLRQYALSGHPTQIFPSSSAGGDFQLEAPRCSSEKGESGETEGPDSSLRKRPSRISRTFFSPHQGDPVEWVEGNREGGTDLRLAQRMSLGGSDTMLKGADTSESGAVIRGNYRLGLSKKSSLFSHQKH | |||
<font color = blue>HVCPECGRGFCQRSDLIKHQRTHTGEKP | |||
YLCPECGRRFSQKASLSIHQRKHSGEKP | |||
YVCRECGRHFRYTSSLTNHKRIHSGERP | |||
FVCQECGRGFRQKIALLLHQRTHLEEKP | |||
FVCPECGRGFCQKASLLQHQSSHTGERP | |||
FLCLECGRSFRQQSLLLSHQVTHSGEKP | |||
YVCAECGHSFRQKVTLIRHQRTHTGEKP | |||
YLCPQCGRGFSQKVTLIGHQRTHTGEKP | |||
YLCPDCGRGFGQKVTLIRHQRTHTGEKP | |||
YLCPKCGRAFGFKSLLTRHQRTHSEEEL | |||
YVDRVCGQGLGQKSHLISDQRTHSGEKP | |||
CICDECGRGFGFKSALIRHQRTHSGEKP | |||
YVCRECGRGFSQKSHLHRHRRTKSG</font>HQL LPQEVF* 0 | |||
>ZNF596_homSap Homo sapiens (human) 504 aa KRAB, no early zinc finger but array | |||
0 MPSP 0 | |||
0 DS<font color = green>MTFEDIIVDFTQEEWALLDTSQRKLFQDVMLENISHLVSI 1 | |||
2 GKQLCKSVVLSQLEQVEKLST</font>QRISLLQ 1 | |||
2 GREVGIKHQEIPFIQHIYQKGTSTISTM 0 | |||
0 RSHTQEDPFLCNDLGEDFTQHIALTQNVITYMRTKHFVSKKFGKIFSDWLSFNQHKEIHTKCKSYGSHLFDYAFIQNSALRPHSVTHTREIT | |||
<font color = blue>LECRVCGKTFSKNSNLRRHEMIHTGEKP</font> | |||
<font color = blue>HGCHLCGKAFTHCSDLRKHERTHTGEKP</font> | |||
<font color = blue>YGCHLCGKAFSKSSNLRRHEMIHTREKA</font> | |||
<font color = blue>QICHLCGKAFTHCSDLRKHERTHLGDKP</font> | |||
<font color = blue>YGCLLCGKAFSKCSYLRQHERTHNGEKP</font> | |||
<font color = blue>YECHLCGKAFSHCSHLRQHERSHNGEKP</font> | |||
<font color = blue>HGCHLCGKAFTESSVLKRHERIHTGEKP</font> | |||
<font color = blue>YECHVCGKAFTESSDLRRHERTHTGEKP</font> | |||
<font color = blue>YECHLCGKAFNHSSVLRRHERTHTGEKP</font> | |||
<font color = blue>YECNICGKAFNRSYNFRLHRRVHTGEKP</font> | |||
<font color = blue>YVCPLCGKAFSKFFNLRQHERTHT</font>KKAMNM* 0 | |||
>GAS8_homSap Homo sapiens (human) synteny marker centromeric to PRDM7 in placentals | |||
0 M 0 | |||
0 APKKKGKKGKAKGTPIVDGLAPEDMSKEQ 0 | |||
0 VEEHVSRIREELDREREERNYFQLERDKIHTFWEITRRQLEEKKAELRNKDREMEEAEERHQVEIK 0 | |||
0 VYKQKVKHLLYEHQNNLTEMKAEGTVVMKLAQKEHRIQESVLRKDMRALKVELKEQELASEVVVKNLRL 0 | |||
0 KHTEEITRMRNDFERQVR 1 | |||
2 EIEAKYDKKMKMLRDELDLRRKTELHEVEERKNGQIHTLMQRHEEAFTDIKNYYNDITLNNLALINSLK 0 | |||
0 EQMEDMRKKEDHLEREMAEVSGQNKRLADPLQKAREEMSEMQKQLANYERDKQILL 0 | |||
0 CTKARLKVREKELKDLQWEHEVLEQRFTK 0 | |||
0 VQQERDELYRKFTAAIQEVQQKTGFKNLVLERKLQALSAAVEKKEVQFNEVLAASNLDPAALTLVSRKLE 0 | |||
0 DVLESKNSTIKDLQYELAQVCK 0 | |||
0 AHNDLLRTYEAKLLAFGIPLDNVGFKPLETAVIGQTLGQGPAGLVGTPT | |||
>CDH12_homSap Homo sapiens (human) synteny marker chr 5 794 aa | |||
MLTRNCLSLLLWVLFDGGLLTPLQPQPQQTLATEPRENVIHLPGQRSHFQRVKRGWVWNQFFVLEEYVGSEPQYVGKLHSDLDKGEGTVKYTLSGDGAGTVFTIDETTGDIHAIRSLDRE | |||
EKPFYTLRAQAVDIETRKPLEPESEFIIKVQDINDNEPKFLDGPYVATVPEMSPVGAYVLQVKATDADDPTYGNSARVVYSILQGQPYFSIDPKTGVIRTALPNMDREVKEQYQVLIQAK | |||
DMGGQLGGLAGTTIVNITLTDVNDNPPRFPKSIFHLKVPESSPIGSAIGRIRAVDPDFGQNAEIEYNIVPGDGGNLFDIVTDEDTQEGVIKLKKPLDFETKKAYTFKVEASNLHLDHRFH | |||
SAGPFKDTATVKISVLDVDEPPVFSKPLYTMEVYEDTPVGTIIGAVTAQDLDVGSSAVRYFIDWKSDGDSYFTIDGNEGTIATNELLDRESTAQYNFSIIASKVSNPLLTSKVNILINVL | |||
DVNEFPPEISVPYETAVCENAKPGQIIQIVSAADRDLSPAGQQFSFRLSPEAAIKPNFTVRDFRNNTAGIETRRNGYSRRQQELYFLPVVIEDSSYPVQSSTNTMTIRVCRCDSDGTILS | |||
CNVEAIFLPVGLSTGALIAILLCIVILLAIVVLYVALRRQKKKDTLMTSKEDIRDNVIHYDDEGGGEEDTQAFDIGALRNPKVIEENKIRRDIKPDSLCLPRQRPPMEDNTDIRDFIHQR | |||
LQENDVDPTAPPYDSLATYAYEGSGSVAESLSSIDSLTTEADQDYDYLTDWGPRFKVLADMFGEEESYNPDKVT* | |||
>CDH10 | |||
MTIHQFLLLFLFWVCLPHFCSPEIMFRRTPVPQQRILSSRVPRSDGKILHRQKRGWMWNQFFLLEEYTGSDYQYVGKLHSDQDKGDGSLKYILSGDGAGTLFIIDEKTGDIHATRRIDRE | |||
EKAFYTLRAQAINRRTLRPVEPESEFVIKIHDINDNEPTFPEEIYTASVPEMSVVGTSVVQVTATDADDPSYGNSARVIYSILQGQPYFSVEPETGIIRTALPNMNRENREQYQVVIQAK | |||
DMGGQMGGLSGTTTVNITLTDVNDNPPRFPQNTIHLRVLESSPVGTAIGSVKATDADTGKNAEVEYRIIDGDGTDMFDIVTEKDTQEGIITVKKPLDYESRRLYTLKVEAENTHVDPRFY | |||
YLGPFKDTTIVKISIEDVDEPPVFSRSSYLFEVHEDIEVGTIIGTVMARDPDSISSPIRFSLDRHTDLDRIFNIHSGNGSLYTSKPLDRELSQWHNLTVIAAEINNPKETTRVAVFVRIL | |||
DVNDNAPQFAVFYDTFVCENARPGQLIQTISAVDKDDPLGGQKFFFSLAAVNPNFTVQDNEDNTARILTRKNGFNRHEISTYLLPVVISDNDYPIQSSTGTLTIRVCACDSQGNMQSCSA | |||
EALLLPAGLSTGALIAILLCIIILLVIVVLFAALKRQRKKEPLILSKEDIRDNIVSYNDEGGGEEDTQAFDIGTLRNPAAIEEKKLRRDIIPETLFIPRRTPTAPDNTDVRDFINERLKE | |||
HDLDPTAPPYDSLATYAYEGNDSIAESLSSLESGTTEGDQNYDYLREWGPRFNKLAEMYGGGESDKDS | |||
>PRDM7_musMus1 Mus musculus genomic strain | |||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTQNSHLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSNLIKHQRTHTGEKP | |||
YVCRECGWGFTQKSDLIQHQRTHTREKp | |||
>PRDM7_musMus2 Mus musculus strain WSB/EiJ GU183911 and EU719625 missing a repeat | |||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTQNSHLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSNLIKHQRTHTGEKP | |||
YVCRECGWGFTQKSDLIQHQRTHTREK | |||
>PRDM7_musMus3 Mus musculus strain MOLF/EiJ GU183913 | |||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTEKSSLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSNLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
>PRDM7_musMus4 Mus musculus strain PWD/PhJ GU183912 = PWD/Ph FJ212287 | |||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTEKSSLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
>PRDM7_musMus5 Mus musculus strain CAST/EiJ GU183909 | |||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTEKSSLIKHQRTHTGEKP | |||
YVCRECGWGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSSLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKPa | |||
YVCRECGWGFTQKSNLIKHQRTHTGEKP | |||
YVCRECGWGFTQKSDLIQHQRTHTREK* | |||
>PRDM7_musMus6 Mus musculus strain C57BL10.F HQ704390 | |||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTQNSHLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTVKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTQNSHLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTQNSHLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSNLIKHQRTHTGEKP | |||
YVCRECGWGFTQKSDLIQHQRTHTREK | |||
>PRDM7_musMol Mus musculus molossinus GU216230 | |||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTEKSSLIKHQRTHTGEKP | |||
YVCRECGWGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSSLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGWGFTQKSNLIKHQRTHTGEKP | |||
YVCRECGWGFTQKSDLIQHQRTHTREK* | |||
>PRDM9_musCas Mus musculus castaneus | |||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSVLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTARSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTEKSSLIKHQRTHTGEKP | |||
YVCRECGWGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSSLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIQHQRTHTGEKP | |||
YVCRECGWGFTQKSNLIKHQRTHTGEKP | |||
YVCRECGWGFTQKSDLIQHQRTHTREKp | |||
>PRDM9_musPah Mus pahari | |||
SIERQCGQYFSDKSNVNEHQRTHTGEKP | |||
YVCRECGRGFTQKSNLITHQRTHTGEKP | |||
YVCRECGRGFTGKSPLIRHQRTHTGEKP | |||
YVCRECGRGFTQKSNLITHQRTHTGEKP | |||
YVCRECGRGFTGKSPLIRHQRTHTGEKP | |||
YVCRECGRGFTQKSHLIKHQRTHTGEKP | |||
YVCRECGRGFTEKSNLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSPLIRHQRTHTGEKP | |||
YVCTECGRGFTQKSNLITHQRTNTGEKP | |||
>PRDM9_musMac Mus macedonicus | |||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTVKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTVKSHLTQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSHLIKHQRTHTGEKP | |||
YVCRECGRGFTQKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIKHQRTHTGEKP | |||
YVCRECGRGFTQNSHLTQHQRTHTGEKS | |||
YVCRECGWGFKQKSDLIQHQRTHTREKp | |||
>PRDM9_musSpi Mus spicilegus | |||
SIERQCGQYFSDKSNVNEHQKTHTGEKP | |||
YVCRECGRGFTQKSNLIQHQRTHTGEK | |||
YVCRECGRGFTQKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTVKSHLTQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLIKHQRTHTGEKP | |||
YVCRECGRGFTAKSHLTQHQRTHTGEKP | |||
YVCRECGRGFTQKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTAKSNLIKHQRTHTGEKP | |||
YVCRECGRGFTQNSHLTQHQRTHTGEKS | |||
YVCRECGWGFKQKSDLIQHQRTHTREKp | |||
>PRDM9_merUng Meriones unguiculatus | |||
GTGRECGQCFSDKSNVSEHQRTHTGEKP | |||
YVCRECGRGFMQRSNLISHQRTHTGEKP | |||
YVCRECGRGFMQRSNLISHQRTHTGEKP | |||
YVCRECGRGFTVKSVLISHQRTHTGEKP | |||
YVCRECGRGFTVKPHLISHQRTHTGEKP | |||
HVCRECGRGFTQRSNLIRHQRTHTGEKP | |||
YVCRECGRGFTVKPHLISHQRTHTGEKP | |||
YVCRECGRGFTVKPHLISHQRTHTGEKP | |||
YVCRECGRGFTVKSVLISHQRTHTGEKP | |||
YVCRECGRGFTVKSVLIRHQRTHTGEKP | |||
YVCRECRRGFTQRSTLIRHQRTHTGEKP | |||
HVCRECGRGFTRGSHLLRHQRTHTGEVL | |||
>PRDM9_micAgr Microtus agrestis | |||
RVGGERGQCFSDKSNVNEHQRTHTGEKP | |||
YVCRECGRGFTRKSNLNVHQRTHTGEKP | |||
YVCRECGRGFTRKALLISHQRTHTGEKP | |||
YVCRECGRGFTQKALLISHQRTHTGEKP | |||
YVCRECGRGFTQKSYLILHQRTHTGEKP | |||
YVCRECGRGFTGKSNLNVHQRTHTGEKP | |||
YVCRECGRGFTQKSYLILHQRTHTGEKP | |||
YVCRECGRGFTGKSLLIRHQRTHTGEKP | |||
YVCRECGRGFTQKSYPILHQRTHTGEKp | |||
>PRDM9_arvTer Arvicola terrestris | |||
RVEGECGQCFNDKSNVNERQRTHTGEKP | |||
YVCRECGRGFTRKSVLILHQRTHTGEKP | |||
YVCRECGRGFTQKSVLINHQRTHTGEKP | |||
YVCRECGRGFTQKSHLIFHQRTHTGEKP | |||
YVCRECGRGFTQKSHLILHQRTHTGEKP | |||
YVCRECGRGFTWKSVLILHQRTHTGERP | |||
YVCRECGRGFTRKSHLILHQRTHTGEKP | |||
YVCRECGRGFTQKSHLILHQRTHTGEKP | |||
YVCRECGRGFTRKSVLILHQRTHTGEKP | |||
YVCRECGRGFTRKSVLINHQRTHTGEKp | |||
>PRDM9_perPol Peromyscus polionotus | |||
RIETECGQRFSDKSNVNESQRTHSEEKP | |||
YVCRECGQGFIQKSVLICHQRTHTGEKP | |||
YVCRECGQGFTWKSHLIRHQRTHTGEKP | |||
YVCRECGKGFIRKSHLICHQRTHTGEKP | |||
YVCRECGQGFIQKSHLICHQRTHTGEKP | |||
YVCRECGQGFTQKSVLICHQRTHTGEKP | |||
YVCRECGQGFIRKSYLICHQRTHTGEKP | |||
YVCRECGKGFTWKSVLIRHQRTHTVEKp | |||
>PRDM9_perLeu Peromyscus leucopus | |||
RIETECGQRFSDKSNANESQRTHSEEKP | |||
YVCRECGQGFTRKSYLICHQRTHTGEKP | |||
YVCRECGQGFIQKSVLIRHQRTHTGEKP | |||
YVCRECGQGFTRKSYLICHQRTHTGEKP | |||
YVCRECGQGFIQKSVLIRHQRTHTGEKP | |||
YVCRECGQGFTWKSVLICHQRTHTGEKP | |||
YVCRECGQGFTRKSYLICHQRTHTGEKP | |||
YVCRECGQGFTWKSHLIRHQRTHTGEKP | |||
YVCRECGQGFTRKSYLICHQRTHTGEKP | |||
YVCRECGQGFIQKSHLICHQRTHTGEKP | |||
YVCRECGQGFTRKSYLICHQRTHTGEKP | |||
YVCRECGQGFTWKSVLIRHQRTHTAEKp | |||
>PRDM9_perMan Peromyscus maniculatus | |||
RTETECGQHFSDKSNANESQRTHSEEKP | |||
YVCRECGQGFTWKSVLIRHQRTHTGEKP | |||
YVCRECGQGFTWKSVLICHQRTHTGEKP | |||
YVCRECGQGFTWKSVLICHQRTHTGEKP | |||
YVCRECGQGFIQKSHLIRHQRTHTGEKP | |||
YVCRECGQGFIRKSHLICHQRTHTGEKP | |||
YVCRECGQGFAQKSVLIYHQRTHTGEKP | |||
YVCRECGQGFTRKSHLICHQRTHTGEKP | |||
YVCRECGQGFAQKSVLICHQRTHTGEKP | |||
YVCRECGQGFTWKSVLICHQRTHTGEKP | |||
YVCRECGQGFIQKSHLIRHQRTHTGEKP | |||
YVCRECGQGFIQKSHLIRHQRTHTGEKp | |||
>PRDM9_apoSyl Apodemus sylvaticus | |||
RVERQRGQCFSDKSNVSERQGTHTGEKP | |||
CVCRECGRGFTQKSHLNRHQRTHTGEKP | |||
HVCRECGRGFTQKSHLNRHQRTHTGEKP | |||
HVCRECGRGFTLKSNLNRHQRTHTGEKP | |||
CVCRECGRAFTQKSDLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSNLNQHQRTHTGEKP | |||
YVCRECGRGFTRKSLLIQHQRTHTGEKP | |||
YVCRECGRGFTQKSDLNRHQRTHTGEKP | |||
YVCRECGRGLTQKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTLKSDLIQHQRTHTGEKP | |||
YVCRECGRGFTRKSDLNRHQRTHTGEKP | |||
YVCRECGRGFTQKSNLIQHQRTHTGEKP | |||
YVCRECGRGFTLKSDLIQHQRTHTGEKP | |||
YVCRECGRGFTRKSDLNRHQRTHTGEKp | |||
>PRDM7_ratNor Rattus norvegicus | |||
RIERQCGQCFSDKSNVSEHQRTHTGEKP | |||
YICRECGRGFSQKSDLIKHQRTHTEEKP | |||
YICRECGRGFTQKSDLIKHQRTHTEEKP | |||
YICRECGRGFTQKSDLIKHQRTHTGEKP | |||
YICRECGRGFTQKSDLIKHQRTHTEEKP | |||
YICRECGRGFTQKSSLIRHQRTHTGEKP | |||
YICRECGLGFTQKSNLIRHLRTHTGEKP | |||
YICRECGLGFTRKSNLIQHQRTHTGEKP | |||
YICRECGQGLTWKSSLIQHQRTHTGEKP | |||
YICRECGRGFTWKSSLIQHQRTHTVEKp | |||
=== Online references === | |||
Open [http://www.ncbi.nlm.nih.gov/pubmed/22291443,22162947,22028627,22102853,21697099,22006216,2175015,21460839,21775986,21564555,21330546,21604305,21698098,20385822,21366357,21334090,21388701,19349985,21159001,21343182,20044539,20818382,21170346,20150474,20334833,20981099,20877321,20044541,20210982,20044538,20948797,20961408,19168450,19074312,19997497,20041164,19150836,18941885,17660532,17916234,17032681,16582607,10656784,19165926,18325330,15705809,20961408,20346131,20478078,19261184,18650392,16292313,19175294 53 recent PubMed abstracts] on PRDM9 and related issues. Or use the reverse chronological list below to get free full text for individual articles when that is available: | |||
[http://www.ncbi.nlm.nih.gov/pubmed/22291443 abs 2012] Sarbajna A major recombination hotspot in the XqYq pseudoautosomal region gives new insight into processing of human gene conversion events. Hum Mol Genet. 2012 Feb 8 | |||
[http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1001211 htm 2011] Ségurel The Case of the Fickle Fingers: How the PRDM9 Zinc Finger Protein Specifies Meiotic Recombination Hotspots in Humans. PLoS Biol 9(12): e1001211. doi:10.1371/journal.pbio.1001211 | |||
[http://gbe.oxfordjournals.org/content/3/614.full htm 2011] Katzman Ongoing GC-Biased Evolution Is Widespread in the Human Genome and Enriched Near Recombination Hot Spots. Genome Biol Evol (2011) 3 614-626 | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3213085/?tool=pubmed htm 2011] Muñoz Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS One. Nov 2011; 6(11): e25498. | |||
[http://genome.cshlp.org/cgi/pmidlookup?view=long&pmid=22006216 htm 2011] Axelsson Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 17 Oct 2011 | |||
[http://dx.plos.org/10.1371/journal.pbio.1001176 htm 2011] Grey Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 2011 Oct;9(10):e1001176. 2011 Oct 18. | |||
[http://www.pnas.org/content/108/30/12378.full.pdf+html?with-ds=yes pdf 2011] Berg Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. PNAS 2011 Jul 26;108(30):12378-83. | |||
[http://onlinelibrary.wiley.com/store/10.1111/j.1747-0285.2011.01135.x/asset/supinfo/CBDD_1135_sm_TableS1-FigS1-4.pdf?v=1&s=81ef430033f1fd8d18bed72183e312564387ad66 pdf 2011] Richon Chemogenetic analysis of human protein methyltransferases. Chem Biol Drug Des. 2011 Aug;78(2):199-210. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/21366357 abs 2011] Campagna Structural Chemistry of the Histone Methyltransferases Cofactor Binding Site. J Chem Inf Model. 2011 Mar 28;51(3):612-23. | |||
[http://genetics.med.harvard.edu/reich/Reich_Lab/Welcome_files/2011_Hinch_Nature.pdf pdf 2011] Hinch The landscape of recombination in African Americans. Nature. 2011 Jul 20. doi: 10.1038/nature10336. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3117304/ pmc 2011] Smagulova Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011 Apr 21;472(7343):375-8. | |||
[http://www.sciencemag.org/content/331/6019/916.full htm 2011] Kaupi Distinct properties of the XY pseudoautosomal region crucial for male mouse meiosis. Science 18 Feb 2011;DOI: 10.1126/science.1195774 | |||
[http://www.ncbi.nlm.nih.gov/pubmed/21604305 abs 2011] Briknarova The PR/SET domain in PRDM4 is preceded by a zinc knuckle. Proteins 2011 Jul;79(7):2341-5. doi: 10.1002/prot.23057. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3117798 pmc 2011] Fledel Variation in human recombination rates and its genetic determinants. PLoS One 2011;6(6):e20321. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/21334090 abs 2011] Neaves Unisexual reproduction among vertebrates. Trends Genet. 2011 Mar;27(3):81-8. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/21388701 abs 2011] Ponting What are the genomic drivers of the rapid evolution of PRDM9? Trends Genetics (2011) 1–7 | |||
[http://nar.oxfordjournals.org/content/early/2011/02/21/nar.gkr048.full htm 2011] Yanover Extensive protein and DNA backbone sampling improves structure-based specificity prediction for C2H2 zinc fingers. Nucleic Acids Res. 2011 Feb 22 | |||
[http://www.tiem.utk.edu/~fubeda/papers/U&W11JEB.pdf pdf 2011] Ubeda Red Queen theory of recombination hotspots. J Evol Biol. 2011 Mar;24(3):541-53. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/20334833 abs 2010] Hochwagen Meiosis: a PRDM9 guide to the hotspots of recombination. Curr Biol. 2010 Mar 23;20(6):R271-4. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2867876 pmc 2010] Tsai Conservation of recombination hotspots in yeast. PNAS 2010 April 27; 107(17): 7847–7852. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/20478078 abs 2010] Klug The discovery of zinc fingers and practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010 Feb;43(1):1-21. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/20818382 abs 2010] Berg PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010 Oct;42(10):859-63. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/20877321 abs 2010] McVean PRDM9 marks the spot. Nat Genet. 2010 Oct;42(10):821-2. | |||
[http://eebweb.arizona.edu/nachman/Suggested%20Papers/Lab%20papers%20fall%202010/Kong_et_al_2010.pdf pdf 2010] Kong Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010 Oct 28;467(7319):1099-103. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2821451 pmc 2010] Parvanov Prdm9 controls activation of mammalian recombination hotspots. Science. 2010 Feb 12;327(5967):835. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2865497 pmc 2010] Lorenz The ancient mammalian KRAB zinc finger gene cluster on human chromosome 8q24.3 BMC Genomics. 2010 Mar 26;11:206. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2872867 pmc 2010] Neale PRDM9 points the zinc finger at meiotic recombination hotspots. Genome Biol. 2010;11(2):104. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2950028 pmc 2010] Sandovici PRDM9 sticks its zinc fingers into recombination hotspots and between species. F1000 Biol Rep. 2010 May 24;2. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2999565 pmc 2010] Billings Patterns of recombination activity on mouse chromosome 11 revealed by high resolution mapping. PLoS One. 2010 Dec 8;5(12):e15340. | |||
[http://www.sciencemag.org/content/327/5967/791.full?rss=1 htm 2010] Cheung Genetic control of hotspots. Science. 2010 Feb 12;327(5967):791-2. | |||
[http://www.med.upenn.edu/timm/documents/Highlyefficientendogenous.pdf pdf 2010] Urnov Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005 Jun 2;435(7042):646-51. | |||
[http://genomebiology.com/2010/11/10/R103 htm 2010] Zheng Detecting sequence polymorphisms associated with meiotic recombination hotspots in the human genome. Genome Biol. 2010;11(10):R103. | |||
[http://www.sciencemag.org/content/327/5967/836.full htm 2010] Baudat PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010 Feb 12;327(5967):836-40. | |||
[http://www.sciencemag.org/content/327/5967/876.full htm 2010] Myers Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010 Feb 12;327(5967):876-9. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2678279/?tool=pubmed pmc 2009] Jeffreys The rise and fall of a human recombination hot spot. Nat Genet 2009 41:625–629. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2631073 pmc 2009] Berglund Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009 Jan 27;7(1):e26. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2667407 pmc 2009] Thomas Evolution of C2H2-zinc finger genes revisited. BMC Evol Biol. 2009 Mar 4;9:51. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2779102 pmc 2009] Oliver Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009 Dec;5(12):e1000753. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2794550 pmc 2009] Thomas Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS One. 2009 Dec 30;4(12):e8505. | |||
[http://www.sciencemag.org/content/323/5912/350.full?rss=1 htm 2009] Willis Origin of species in overdrive. Science. 2009 Jan 16;323(5912):350-1. | |||
[http://www.andrologyjournal.org/cgi/content/full/30/4/426 htm 2009] Irie Single-nucleotide polymorphisms of the PRDM9 (MEISETZ) gene in patients with nonobstructive azoospermia. J Androl. 2009 Jul-Aug;30(4):426-31. | |||
[http://www.sciencemag.org/content/323/5912/373.full htm 2009] Mihola A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009 Jan 16;323(5912):373-5. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/18286240 abs 2008] Brayer The protein-binding potential of C2H2 zinc finger domains. Cell Biochem Biophys. 2008;51(1):9-19. | |||
[http://www.pnas.org/content/105/30/10471.long htm 2008] Webb Sperm cross-over activity in regions of the human genome showing extreme breakdown of marker association. PNAS 2008 105:10471–10476 | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2346554 pmc 2008] Duret The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008 May 9;4(5):e1000071. | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2346554 pmc 2008] Duret The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008 May 9;4(5):e1000071. | ||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2593767 pmc 2008] Miyamoto Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet. 2008 Nov-Dec;25(11-12):553-7. | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2593767 pmc 2008] Miyamoto Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet. 2008 Nov-Dec;25(11-12):553-7. | ||
[http://bioinfo.hanyang.ac.kr/ZIFIBI/frameset.php htm 2008] Cho Prediction of DNA binding sites for zinc finger proteins. BBRC 2008 May 9;369(3):845-8. | [http://bioinfo.hanyang.ac.kr/ZIFIBI/frameset.php htm 2008] Cho Prediction of DNA binding sites for zinc finger proteins. BBRC 2008 May 9;369(3):845-8. | ||
[http://www.ncbi.nlm.nih.gov/pubmed/19165926 abs 2008] Myers A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008 Sep;40(9):1124-9. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1817654 pmc 2007] Coop Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007 Mar 9;3(3):e35. | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1817654 pmc 2007] Coop Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007 Mar 9;3(3):e35. | ||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2082429 pmc 2007] Fumasoni Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007 Oct 4;7:187. | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2082429 pmc 2007] Fumasoni Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007 Oct 4;7:187. | ||
[http://www.genetics.org/content/177/2/881.full htm 2007] Gay Estimating meiotic gene conversion rates from population genetic data. Genetics. 2007 Oct;177(2):881-94. | |||
[http://www.genetics.iastate.edu/Phillips_Dernburg_2006.pdf pdf 2006] Phillips A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis. Dev Cell. 2006 Dec;11(6):817-29. | [http://www.genetics.iastate.edu/Phillips_Dernburg_2006.pdf pdf 2006] Phillips A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis. Dev Cell. 2006 Dec;11(6):817-29. | ||
[http://bioinformatics.oxfordjournals.org/content/22/23/2841.long htm 2006] Birtle Meisetz and the birth of the KRAB motif. Bioinformatics. 2006 Dec 1;22(23):2841-5. | [http://bioinformatics.oxfordjournals.org/content/22/23/2841.long htm 2006] Birtle Meisetz and the birth of the KRAB motif. Bioinformatics. 2006 Dec 1;22(23):2841-5. | ||
[http://www.landesbioscience.com/journals/cc/hayashiCC5-6.pdf pdf 2006] Hayashi Meisetz, a novel histone tri-methyltransferase, regulates meiosis-specific epigenesis. Cell Cycle. 2006 Mar;5(6):615-20. | [http://www.landesbioscience.com/journals/cc/hayashiCC5-6.pdf pdf 2006] Hayashi Meisetz, a novel histone tri-methyltransferase, regulates meiosis-specific epigenesis. Cell Cycle. 2006 Mar;5(6):615-20. | ||
[http://www.img.cas.cz/journalclub/fulltexts/07_10_18_odd21_28.pdf pdf 2005] Hayashi A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005 Nov 17;438(7066):374-8. | [http://www.img.cas.cz/journalclub/fulltexts/07_10_18_odd21_28.pdf pdf 2005] Hayashi A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005 Nov 17;438(7066):374-8. | ||
[http://www.sciencemag.org/content/308/5718/107.full htm 2005] Winckler Comparison of Fine-Scale Recombination Rates in Humans and Chimpanzees. Science 2005 2005 Apr 1;308(5718):107-11. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/10656784 abs 2000] Laity DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J Mol Biol. 2000 Jan 28;295(4):719-27. | [http://www.ncbi.nlm.nih.gov/pubmed/10656784 abs 2000] Laity DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J Mol Biol. 2000 Jan 28;295(4):719-27. | ||
=== Article author === | |||
[[Image:Author.jpg|left]] | |||
I researched this article in its entirety in April and July-August of 2011, paying as little attention as possible to the previous studies above, which are excellent on meiosis but completely clueless on comparative genomics. This is a moderately difficult topic as human genes go, so the overall annotation is still being revised periodically into 2012 as better quality genomes become available. A change in one section places others in need of revision, not always promptly attended to. | |||
Although copyrighted, all the information here is in the public domain and can be used by anyone without additional permissions if properly sourced; however if data, figures or original observations are taken for a peer-reviewed scientific publication, it might be appropriate (after consultation early on) to include me among secondary co-authors. | |||
Rather than make article edits yourself, please contact me by email with clarifications, corrections or additions to the content so I can make edits while maintaining a consistent approach. For broader disagreements or different interests, a better option is to register at the UCSC genomeWiki site and create your own page within the comparative genomics category. | |||
This is just a scientific research article on a vertebrate gene family, not a counseling resource for personal genomics nor medical advice on infertility -- thanks in advance for not sending inappropriate email. Note technical terms from genetics and molecular biology are not explained when keywords have a satisfactory treatment at wikipedia or in undergraduate genetics texts. | |||
My last dozen published research papers in PNAS, Nature, Science etc can be found [http://www.ncbi.nlm.nih.gov/pubmed/21709235,20164927,19020620,18464734,18266766,17984227,18085818,17975064,17322288,15608236,12045153,22012981 here]. Watch for 4 additional comparative genomics paper to appear in 2012. I've also written over a [[:Category:Comparative_Genomics|thousand pages of comparative genomics]] for other human genes, authored the original user manual to the UCSC human genome browser and in 1999 an advanced tutorial on metazoan genome annotation still [http://www.mad-cow.org/00/annotation_tutorial.html widely available online]. I thank the UCSC Genomics Group (Hiram Clawson, Brian Raney) for software and manuscript resources, Evim Foundation for logistical support, and the Sperling Foundation for financial support under project grant 2011.GNTCS.004. | |||
<br clear=all> | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 15:21, 16 September 2015
See also: All about PRDM11
Updates: To help readers locate fixes, additions and other news as the article grows longer, significant additions will be noted here in reverse chronological order linked into their spot in the article. 04 Feb 11: improved cow and sheep assemblies have six PRDM7/9 genes and a functional chrX copy with 21 zinc fingers. 15 Dec 11: wake up folks: that gene in dog and mice is PRDM7 not PRDM9 11 Dec 11: added a whole new page on PRDM11 -- the closest match in non-mammalian amniotes to the PR(SET) methylation domain of PRDM7/9 30 Oct 11: added 6 new fragmentary Carnivora PRDM7 sequences that -- like dog -- have inactive terminal exons. 12 Sep 11: started new section on origin of species, sex chromosome co-evolution etc. 11 Sep 11: re-edited first 20 pages for glitches, redundancy and inconsistencies. 09 Sep 11: added sections on chained meiosis in platypus and non-PAR PRDM9 gene conversion sites on human chrY. 07 Sep 11: transversional second block of human PRDM9 recognizes hotspots. 31 Aug 11: the first 523 amino acids of primate PRDM9 are not evolving at an anomalous rate. 28 Aug 11: partial distal pseudogenization of PRDM7 in some catarrhines. 22 Aug 11: re-wrote section proving mouse PRDM9 is really PRDM7; re-analyzed historic species barrier paper. 22 Aug 11: improved mouse PAR region depiction and updated expression data -- retinal transcripts suggest a multi-functional protein. 19 Aug 11: showed odd TGEKL cap region of first human repeat was already present in Denisova and pygmy. 18 Aug 11: fixed timing of the primate PRDM7 duplication creating PRDM9 relative to marmoset, tarsier, lemurs and tree shrew.
Introduction
PRDM9 is a gene on human chromosome 5 with a very peculiar history. Its primary function -- after many false starts -- has only recently become clear: scanning the genome with its terminal zinc finger array to locate and mark recombination hotspots with its histone methylase where its transcription factor binding domain can direct additional proteins to initiate the double stranded breaks needed for meiosis. Some level of recombination between homologous chromosomes is essential to proper alignment and separation into daughter cells as well as for bringing favorable alleles onto the same haplotype for adaptive evolution.
This reaches criticality in placental mammal sex chromosomes which are limited in homologous alignability to short pseudoautosomal regions (PAR). Here in male meiosis, a recognizable sequence site must be found for the double stranded break with only tens of kilobases available in mouse, the most favorable experimental situation. However two large gaps remain in the most recent mouse assembly used (July 07) telomeric to the single known PAR hotspot (a situation not improved in the July 2011 release 37.2) nor fixed by Illumina reads. This region likely consists entirely of sequence categorized as simple repeats, meaning that as one hotspot is erased by gene conversion, similar ones remain available in the region, mitigating the need for adaptation in PRDM7/9.
Humans are unique in having a second PAR of size 330 kb on distal Xq which contains five genes, acquired recently by chrY via LINE-mediated illegitimate recombination. Shrinkage of the larger PAR1 (currently 2.7 Mb, 24 genes) since lemur divergence (a process at much longer time scales driven by strata-creating inversions), reduces the potential number of recombination initiation sites and may correlate with autosomal duplication of PRDM7 around this same time.
Gene conversion also occurs unexpectedly in human at various hotspots between the PARs, notably in the PRKY, VCX/Y, TGIF2LX/Y and IR1 and P1 regions -- as well as intra-chromosomally in the palindromic section of chrY. The PRKY site contains a canonical PRDM9 recognition site, CCCCCCCTTCCCTC. If recombination intermediates resolve as translocation rather than gene conversion, infertile 46,XX males and 46,XY females result.
A protein central to an ancient essential process is usually highly conserved. However this is not the case here at all. Indeed, it proves exceedingly difficult to find a comprehensive set of PRDM9 orthologs even in the 39 sequenced placental mammalian genomes available on 10 Sept 2011, with immense and continuing confusion in the literature caused by independent segmental gene duplications, partial and full pseudogenizations and mix-ups with other composite domain proteins -- all compounded by outright sequencing error in the long terminal zinc finger repeat array.
Although meoitic recombination is a universal feature of eukaryotes, surprisingly the underlying mechanism for it is not. As shown below, the PRDM9 scenario is not directly applicable even to other major placental mammal clades. And neither PRDM9 nor its parent gene PRDM7 have a full-length orthologous counterpart in monotremes, birds, lizards, amphibians or earlier diverging vertebrates. While similar domain combinations have arisen before in other narrow bilateran clades, no evidence connects them to meiosis. Drosophila in particular uses very different gene products; meiosis there does not involve zinc finger proteins.
This puzzling history did not arise from ab initio sequence innovation in post-Cambrian deuterostomes because PRDM7 (the parent of primate PRMD9) arose instead from chimerization events in the mammalian stem involving SSX1, PRDM11 and a ZNF that together provided the six structural domains of the current protein. The parental gene histories are themselves complex, involving still earlier gene duplications and internal tandem repeat expansions -- patterns with numerous precedents in the overall metazoan proteome evolutionary context. Zinc finger proteins are a greatly expanded, often chimeric family within the mammalian lineage whose history may never be fully unravelled.
Rapid evolution of the terminal region of PRDM7/9 occurs at the amino acid level, both in the number of zinc fingers and -- within a given finger -- in the four non-adjacent residues primarily responsible for recognizing a specific dna trinucleotide. This variability is not coincidental to the role in meiosis -- that process tends to destroy its recombination hotspots by biased gene conversion. Since recombination is essential, new hotspots must emerge. The race is then on for PRDM7 or its spun-off PRDM9s to rapidly evolve and define new histone markup sites.
The consequent mutational pattern is quite distinct from those of closely related zinc finger proteins. It may result from error-prone replication slippage in the repeat region but why that mechanism would concentrate non-synonomous change at certain residue positions remains unexplained, along with fine structure details in meiotic data, transcription in non-meiotic retina and the functions of the conserved zinc knuckle and early zinc finger domains.
This rapid evolution can cause breeding incompatibility between populations in the F1 generation (meiosis arrest for lack of cross-overs, notably between chrX and chrY) and thus be central to the process of speciation. However PRDM7/9 cannot provide a universal explanation of either speciation or Haldane's rule even within placentals because the hotspot-defining genes are not in straightforward correspondence.
In effect, initiation of meiotic recombination is rapidly evolving -- and taking speciation and Haldane's rule along with it: each major clade of placentals has evolved a different hotspot recognition system, taking its most extreme form in pecoran ruminants with six PRDM7/9 genes (whose individual roles are not understood but could be quickly worked out in cattle). These differences within placentals follows upon the very different structure and gene content of sex chromosomes between monotremes, marsupials and placentals which in turn are quite different from those of birds, lizards and the amniote ancestor.
Syntenic relationships can help resolve gene duplication events during mammalian evolution. Here the chromosomal gene order TUBB3+ AFG3L1+ GAS8+ has stably existed since the stem amniote some 310 million years ago, with PRDM7- qTer added in placental mammals after marsupial divergence and maintained there since over billions of years of observable branch length time. PRDM9 however is found in many syntenic contexts, depending on timing and positioning of the segmental duplications giving rise to these secondary copies.
From the perspective of evolutionary genomics, PRDM7 is the fundamental gene, not the disparate collection of genes lumped under PRDM9 (even as those have taken over in primate meiois). At different times in different placental clades, PRDM7 spun off segmental duplications of itself to other sites in other chromosomes, probably because of a susceptible location at the extreme q arm of an autosomal chromosome. Because PRDM7 has stayed at its site adjacent to GAS8, it is possible to say unambiguously which of two initially identical copies was the parent gene. Due to this history, the 'PRDM9' genes do not form their own subtree within the overall two-gene tree under phylogenetic algorithms but instead associate more closely with their parental PRDM7 parent.
These paralogous copies -- despite all being called PRDM9 -- are not usefully considered orthologous outside their species clade of origin. Orthology requires (by long-standing definition) vertical descent from a common gene in the last common ancestor of two species. Here primate PRDM9 are descended from a common gene (namely the recent duplicate of PRDM7 in the stem primate ancestor) but 'PRDM9' in afrotheres, pecoran ruminants, rabbits etc arose from different duplications at different times during placental mammal evolution from a rapidly evolving PRDM7 parental gene in those lineages and so -- despite the name -- are not vertically descended from a common primate PRDM9 in their last common ancestor (though all the genes here descend from a single stem placental PRDM7 gene).
In tandem duplications, the parental copy cannot be distinguished but here the second copy was never on an equal syntenic footing. In a small segmental duplication, not all of the upstream regulatory features (which can be a megabase or more away from transcription start) are necessarily carried over. The second copy may be differently expressed from the get-go despite encoding an identical protein.
In mammals, PRDM7 has segmentally duplicated numerous times, in each case to a different site on a different chromosome. While PRDM7 has a moderately long history, primate PRDM9 has none of its own prior to its creation in stem catarrhine. For clarity, the various PRDM7 duplications can be denoted PRDMPpri, PRDM9pec, and PRDM9afr.
Such gene duplicates are sometimes called in-paralogs within a species and co-orthologs across species. However these terms do not reflect the mechanism of duplication and are topologically unstable (depend on the species range included in the gene tree unlike the terms ortholog, paralog and homolog) and have gained limited traction. Synteny creates an asymmetry here because only the parental gene assuredly has all upstream and downstream effectors.
Composite-domain proteins such as PRDM7/9 give rise to a whole new level of terminological muddle as each domain may have its own complex evolutionary history of earlier duplication, shuffling partners, and functional drift. It is very difficult to capture these histories within gene nomenclature.
Comparative genomics of placental mammals
Within Euarchonta, a small segmental duplication encompassing PRDM7 took place in a stem catarrhine primate. The duplicated gene, designated PRDM9 by the human gene nomenclature committee, resides within an altogether new syntenic context -- a cadherin gene complex on an unrelated autosomal chromosome. PRDM9 initially shared meiotic functionality with PRDM7 even as it diverged in amino acid sequence and descended through speciation events into contemporary old world monkeys and great apes. PRDM7 still persists at its original ancestral location (qTer and adjacent to GAS8) but as an overt full-length pseudogene in some lineages (rhesus, gibbon) but not so clearly in others (orangutan). Gene duplication followed by lineage-specific reallocation of functionality is an exceedingly common scenario within metazoan evolution. The timing of gene duplication here means only catarrhine primates have a PRDM9 gene.
Human PRDM7, despite its 3 frameshifts in exon 9 and 10, may still retain N-terminal functions (those not requiring dna recognition by the zinc finger array). PRDM7 is sometimes treated as a conventional gene with splice 'isoforms' despite an internal direct tandem repeat of 88 nucleotides in exon 9 of the human reference sequence that throws off the reading frame and subsequent splice donor to exon 10, which itself has a frameshift (GGGG to GGG) in the second of its three zinc fingers. This is not an anomaly of the reference genome as it is shared across the 1000 Genome Project. The C-terminal pseudogenization of human PRDM7 predated divergence of denisovan, bushman and neanderthal and postdated analogous events in other primates.
Translation into an incorrect reading frame cannot plausibly yield a stable fold, much less zinc fingers that recognize a nucleotide meiotic sequence. Nothing is known about the fate of in vivo transcripts or mature PRDM7 protein but the likeliest scenarios are nonsense-mediated decay or proteolytic trimming of unfolded C-terminal rubbish. Partial pseudogenization is an option for a protein like PRDM7 with multiple quasi-independent domains. Transcripts and alternative splices -- being artifact-rich processes in vertebrates -- do not provide a reliable guide to any stably folded mature protein that may ultimately be produced.
Initially PRDM7 must have shared its role in meiosis with PRDM9 (the sequences and near-upstream regulatory regions being identical), but later PRDM9 took over this role entirely in most primate clades as only it retained the zinc finger array. PRDM7 either retained non-meiotic roles (implied by numerous transcripts in non-meiotic tissue) or acquired other functionality not involving the terminal zinc finger array (but in some species losing all function).
Chimp and gorilla have also lost functionality in the last exon. However the mechanisms of loss -- stop codon in chimp vs exon 10 frameshifts in gorilla -- differ from human. Orangutan PRDM7 may still be functional whereas gibbon is riddled with early stop codons implying total loss. Since pseudogenization is a fairly rapid process, it could not have begun at the time of segmental duplication in stem catarrhine. Instead PRDM7 co-existed with PRDM9 for tens of millions of years, only in the last few million years losing distal functionality independently in various great ape lineages.
Loss of terminal array function took place fairly late in each great ape lineage by independent mutational mechanisms rather than by a shared disabling mutation in a common ancestor. Residual function may or may not be the same in those great apes that have retained the proximal portion of the gene. Partial pseudogenization is structurally acceptable in chimeric domain proteins if domains fold independently and do not significantly interact.
This scenario is strongly supported by alignment of manually curated primate PRDM7/9 sequences up to but not including the final array. If PRDM7 had been inactivated early after duplication, it would have accrued a large number of non-synonymous changes by now, changes oblivious to the conservation status of individual residues in the early domains. The alignment below shows preferential retention of conserved residues, as well as PRDM7 and PRDM9 clustering into distinct gene sub-trees as expected from the time of duplication. Gene conversion can keep duplicated genes synchronized for a time but that mechanism is not applicable over such a time span for non-tandem genes on unrelated chromosomes.
Earlier diverging primates such as new world monkeys and lemurs have a single PRMR7 gene adjacent to GAS8. Although a PRDM9 duplicate could occur within an genome assembly gap, a large multi-exon gap is implausible in multiple assemblies with respectable Sanger trace read coverage, especially given the pedestrian chromosomal location of catarrhine PRDM9. The tarsier situation however is syntenically unclear -- the gene occurs in five separate contigs, mostly single reads. Tree shrew also has unsatisfactory coverage (six exons spread out over two contigs and 3 unassembled traces, a string of Ns in the terminal zinc finger domain, and undeterminable synteny). These scattered contigs could conceivably represent two or more separate genes.
Within Rodentia, the single mouse PRDM7/9 gene lies in a region of confused synteny (relative to human) attributable to chromosomal rearrangements in the rodent clade. The browser screenshot above shows an unrelated region of human chr5 -- not the part that bears human PRDM9 -- as right-syntenic neighbor to mouse PRDM7. The left-syntenic human chr6 segment does not carry human PRDM7 (human chr16). The mouse gene thus has no informative neighbors (not even flanking debris) from GAS8 or cadherins. Similarly the mouse orthologs of GAS8 and cadherins do not contain PRDM7/9 debris.
The rat gene occurs in the same syntenic context as mouse but other rodent genomes (including the new hamster assembly) are too incomplete for synteny to be assessed. Thus the genetic rearrangement taking PRDM7 from its location facing GAS8 to its current position in rodents cannot be accurately timed relative to rodent divergences. The rabbit assembly of Apr 2009 is still quite garbled in the PRDM7-related region and also contains a spurious assembly stutter duplication. The syntenic location is unlike mouse/rat or any other mammal and again there is no debris at the relevant locations in other species. The other lagomorph assembly (pika) is missing its first and last exon so provides no syntenic information.
Thus the history of chromosomal rearrangements of PRDM7-like genes in Glires requires better assemblies in more species before gene rearrangements, gains and losses can be understood. It would be more useful to finish genomes already begun rather than generate thousands of additional fragmentary assemblies as in the 10k vertebrate genome project.
Conceivably, ancestral euarchontoglire PRDM7 duplicated twice to its current locations in rodents and lagomorphs from the ancestral location adjacent to GAS8 with the parental gene later lost twice. However even in this non-parsimonious scenario, the mouse gene cannot legitimately be called PRDM9 because it is still not a strict ortholog of primate PRDM9. A simpler scenario envisions two lineage-specific chromosomal rearrangements of PRDM7 with no gene gain or loss, consistent with Laurasiathere outgroup data indicating a single copy of PRDM7 at euarchontoglire divergence.
Since human PRDM9 arose in early primates as a gene duplication of a much older PRDM7, it was not present at the time of mouse/human divergence (Euarchontoglires). Hence the mouse gene cannot correspond to it. The mouse gene is best taken as a straightforward ortholog of primate PRDM7. Mouse has a great many chromosomal rearrangements relative to ancestral Euarchontoglires, so a translocation here of PRDM7 is unremarkable. The mouse gene is still called PRDM9 in the scientific literature despite the Jan 2002mouse assembly establishing its lack of synteny to the catarrhine-specific gene!
The PRDM7 protein is evolving conservatively in murid rodents though rather rapidly in the amino acids contacting the hotspot dna motif. There are substantial differences between common strains of lab mouse and unsurprisingly these cannot always interbreed (shown below in first six lines as genome strain C57BL/6J, WSB/EiJ, MOLF/EiJ, PWD/PhJ, CAST/EiJ, and C57BL10.F). Note mouse strains vary considerably in the number of zinc finger repeats -- 11 in CAST/EiJ, 12 in the reference genome strain C57BL/6J, 13 in C3H/HeJ and 14 in strain PWD/Ph -- and so in their dna-contacting residues.
The species barrier between B6 and C3H mouse strains is said entirely attributable to a single difference in the number of zinc finger repeats (loss of repeat 10 in B6). As the extra repeat in C3H intercalates a new set of dna-contacting residues, its array recognizes a dna sequence with a 3 bp insertion relative to the recognition sequence of B6 (ie the barrier does not arise from repeat number variation per se). However it has not been established why a mouse heterozygous for separately meiotically functional PRDM7 genes cannot carry out meiosis.
While the species barrier result implicates distal zinc fingers in meiotic recombination, no meiosis occurs in retina, the primary source of (unsought) mouse PRDM7 transcripts at GenBank. Mouse PRDM7 may be multi-functional, with a distinct role regulating gene expression in retina (in the manner of conventional ZNF genes). The proximal block of zinc fingers could be used there for dna recognition, giving rise to dual (or overlapping) selection on the array.
While consistent with meiosis site recognition requiring less than half of the array, a conflict arises because the mysterious mechanism generating mutational variation specifically at the dna-contacting residues for meiosis would be maladaptive for continuing recognition of fixed retinal gene regulatory targets (if any). Here it is imperative to understand what other genes are acting upstream to confine PRDM7 expression to testis and retina. If transcription in retina is specific to one of its many structural components, that could provide insight into its functionality there.
PRDM7_musMus1 SIERQCGQYFSDKSNVNEHQKTHTGEKPYVCRECGRGFTQNSHLIQHQRTHTGEKPYVCRECGRGFTQKSDLIKHQRTHTGEKPYVCRECGRGFTQKSDLIKHQRTHTGEKPYVCRECGRGFTQKSVLIKHQRTHTGEKPYVCRECGRGFTQKSVLIKHQRTHTGEKPYVCRECGRGFTAKSVLIQHQRTHTGEKPYVCRECGRGFTAKSNLIQHQRTHTGEKPYVCRECGRGFTAKSVLIQHQRTHTGEKP-YVCRECGRGFTAKSVLIQHQRTHTGEKPYVCRECGRGFTQKSNLIKHQRTHTGEKPYVCRECGWGFTQKSDLIQHQRTHTREKP-------------------------------------------------------- PRDM7_musMus2 ....................................................................................................................................................................................................................................................................................................A..V..Q.................R......N..K......G...YVCRECGWGFTQKSDLIQHQRTHTREK............................. PRDM7_musMus3 ........................................K.D..K........................V....................................................A..N..Q.....................A.....Q.....................Q..D..K..............................................................................E..S................................................R...A..V.........G........................................................... PRDM7_musMus4 ........................................K.D..K........................V....................................................A..N..Q.....................A.....Q.....................Q..D..K.................................................E..S..K.........................N...........................V....................R...A..V.........G........................................................... PRDM7_musMus5 .......................................AK.N...........................V..Q.................................................A..N..Q.....................E..S....................W......N........................Q..S..K........................N.............A.......W...Q..N..K.................W......D..Q......R..-----------------------------........................................................ PRDM7_musMus6 ...............................................................................................V..V.........................N.H..Q.....................A.....Q.....................QN.H........................Q..D..K.....................Q.....K......................Q..N..K.................W......D..Q......R..-----------------------------........................................................ PRDM7_musMol2 .......................................AK.N...........................V..Q.................................................A..N..Q.....................E..S....................W......N........................Q..S..K........................N.....................W...Q..N..K.................W......D..Q......R..-----------------------------........................................................ PRDM9_musCas .......................................AK.N...........................V..Q.................................................AR.N..Q........................D...........................N........................E..S..K.................W......N.........................Q..S..K.....................A.....Q........................N..K......G...YVCRECGWGFTQKSDLIQHQRTHTREKP............................ PRDM9_musPah ....................R...................K.N..T.....................G..P..R........................N..T.....................G..P..R........................H........................E..N..K.....................Q..P..R.............T.......Q..N..T....N......------------------------------------------------------------------------------------........................................................ PRDM9_musMac ........................................K.D..K.....................V..............................N..Q........................D........................V..H.TQ.....................Q..D..K........................H..K.....................Q..N............................N..K......................N.H.TQ.........S..........K......................................................................... PRDM9_musSpi ........................................K.N............-..............N..Q.....................A...........................V..H.TQ........................D...........................H.T......................Q..............................N..K......................QN.H.T..........S.......W..K...D..Q......R...----------------------------........................................................ PRDM7_musMol1 .......................................AK.N...........................V..Q.................................................A..N..Q.....................E..S....................W......N........................Q..S..K........................N.....................W...Q..N..K.................W......D..Q......R...----------------------------........................................................ PRDM9_merUng GTG.E...C.......S...R.................M.R.N..S....................M.R.N..S.....................V..V..S.....................V.PH..S..........H...........R.N..R.....................V.PH..S.....................V.PH..S.....................V.....S......................V.....R................R.....R.T..R..........H......R...RG.H.LR......G.VL........................................................ PRDM9_micAgr RVGGER..C...........R..................RK.N.NV.....................R.AL..S.......................AL..S........................Y..L.....................G..N.NV.....................Q..Y..L.....................G..L..R.....................Q..YP.L...........------------------------------------------------------------------------------------........................................................ PRDM9_arvTer RV.GE...C.N.......R.R..................RK.V..L........................V..N........................H..F........................H..L.....................W.....L........R............R..H..L.....................Q..H..L.....................R.....L......................R.....N..........--------------------------------------------------------........................................................ PRDM9_perPol R..TE...R.........S.R..SE..........Q..I.K.V..C.................Q...W..H..R.................K..IR..H..C.................Q..I...H..C.................Q.........C.................Q..IR..Y..C.................K...W..V..R......V...----------------------------.------------------------------------------------------------------------------------........................................................ PRDM9_perLeu R..TE...R......A..S.R..SE..........Q...RK.Y..C.................Q..I...V..R.................Q...R..Y..C.................Q..I......R.................Q...W.....C.................Q...R..Y..C.................Q...W..H..R.................Q...R..Y..C..................Q..IQ..H..C.................Q...R..Y..C.................Q...W..V..R......A........................................................... PRDM9_perMan RT.TE...H......A..S.R..SE..........Q...WK.V..R.................Q...W..V..C.................Q...W..V..C.................Q..I...H..R.................Q..IR..H..C.................Q..AQ.....Y.................Q...R..H..C.................Q..AQ.....C..................Q...W.....C.................Q..I...H..R.................Q..I...H..R......G........................................................... PRDM9_apoSyl RV...R..C.......S.R.G.......C...........K...NR..........H.............H.NR..........H..........L..N.NR..........C.......A.....D..Q........................N.NQ.....................R..L........................Q..D.NR...................L.Q..N.........................L..D........................R..D.NR.................R......N.........G...YVCRECGRGFTLKSDLIQHQRTHTGEKPYVCRECGRGFTRKSDLNRHQRTHTGEKP PRDM7_ratNor R.......C.......S...R........I........S.K.D..K......E....I......................E....I...........................I............D.........E....I............S..R...........I.....L...Q..N..R.L.........I.....L...R.................I.....Q.L.W..S...............I.........W..S.........V...--------------------------------------------------------........................................................
Laurasiatheres have a quite different history of gene duplication. Many clades simply retain the ancestral condition of a single PRDM7 gene adjacent to GAS8. Vampire bat (but not brown bat) has an additional segmental duplication to a novel location that is today a pseudogene. Insectivores, perissodactyls and early-diverging artiodactyls (alpaca, pig, dolphin) have a single PRDM7 gene in ancestral syntenic location (when determinable), though some genes have too few zinc fingers to define genome-specific hotspots in the manner of primates.
The dog reference genome inexplicably has a PRDM7 full-length pseudogene yet no PRDM9-like gene duplication despite a rather complete assembly, and possibly stable recombination hotspots according to a new low resolution study. This is not an inbreeding artifact because five other species of canids share a frameshift within the early zinc finger of exon 7. Red fox (the outgroup) lacks this frameshift as well as a later shared frameshift in the third terminal zinc finger (thus timing them on the canid gene tree) but has an exon 7 frameshift of its own.
Here it should be stressed that partial pseudogenization can only be ruled out in chimeric domain proteins by sequencing the entire gene, not done here. However based on several early inactivating mutations in dog, these canid PRDM7 are unlikely to be even partially functional. Thus some other mechanism must suffice for initiation of meiotic recombination in canids. Recall PRDM9 in humans explains only 40% of the events so a second mechanism (not necessarily that of canids) seem operative there too.
PRDM7 from gray fox, the ultimate canid outgroup, unfortunately has not been sequenced and may still be functional. The alignment of exon 7 in 31 Laurasiatheres below shows rather few substitutions relative to the outgroup, establishing that pseudogenization is relatively recent. Thus PRDM7 was likely lost in canids around 8 myr ago and so was presumably functional during the 40 myr since divergence.
However it is not so clear that PRDM7 is functional (in the sense of marking up meiotic recombination sites with a terminal zinc finger array) in the next outgroup (mink + bear). The first two repeats in the three species available have lost key conserved repeat residues for zinc binding and may be dysfunctional. Panda has an additional 3 repeats but this is still not sufficient to provide a recognition system of sufficient specificity as seen in mouse and human (or for that matter cat or bat). The ends of the mink and ferret PRDM7 genes are not satisfactorily covered by contigs so the number of zinc fingers there cannot be determined.
Carnivores -- but not bats or horses -- have an intervening cadherin gene (CAD1) between GAS8 and PRDM7 which should not be confused with the weakly related cadherins (CAD10 and CAD12: 36% identity to CAD1) flanking primate PRDM9. This rare genomic event does not represent the ancestral state but is unfortunately too restricted in distribution to resolve the status of Pegasoferae.
Given the well-established phylogeny of canids within carnivores, the first indel in the alignment below (a deletion of 4 amino acids) is perplexing because it unifies mink with canids which do not share a common ancestor to the exclusion of other carnivores according to the species tree. The tree topology may be slightly wrong (despite overwhelming statistical support), with mink sistered to canids, not bears. An early mutation followed by lineage-sorting seems implausible as the heterozygous state would have had to persisted 6 myr until mink/bear divergence. Convergent evolution -- an independent deletion in mink of the same length at the same site (respectively an independent insertion in bear) -- seems equally implausible unless special predisposing local dna attributes exist. Sequencing error can be ruled out since two mink, bear and panda, and seven canids provide a consistency check. The second indel -- a single amino acid insertion (serine) in ancestral canid -- is fully consistent with the gene tree. Given that this portion of exon 7 seems under little selectional constrain, neither indel is likely to have functional significance.
Difference alignment of exon 7 relative to dog shows relatively few substitutions given the rapid overall rate of evolution of the inter-domain region: PRDM7_canFam EPNPEIHPCPSCSLAFSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTT----CEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR PRDM7_canLup ................................................................................................................................................ PRDM7_canAur ................................................................................................................................................ PRDM7_lycPic ...................................................................S..........................................................................M. PRDM7_canMes ....................................Q........................................................................................................... PRDM7_speVen ....................................-................................................R..................................S....................... PRDM7_vulVul ......Y.......S........................I....Q....................C........................K..................................................... PRDM7_neoVis ..K.........T.............KC...........AG...Q.E.....E..H.........S...K.....V..S....LE......N....P.......G....Y..M.E..S.T.-......E.E...M....S.M.. PRDM7_musPut ..K.........T..............R...........AG...Q.E.....E..H.........N...K....DV..S....LE.....KN....PI..E...G....Y....E....T.-......E.....M....S.... PRDM7_ailMel ..K................................S.K.AS...QQE.....H...........H....K.....V.......L.............S......RSSTV...M.E......-......E.....M....SG... PRDM7_felCat ..Q.D..R.................V.CK.S..S..Q..A.K..Q.EN....D...........HS...K..C..V...SR..L...K...............MGSSRV...M.E.G..M.-.N..S.......M....S..V. PRDM7_equCab ..KL.....................V.R........GT.A.N.LQ.E..S..D......-....HS.K.K.HS..V...S...L.K......P....Y.P...MENFRMR.R.ME.K..I.-R.V.........LEMR.S.NV. PRDM9_pteVam ..K...............R......MKRS....S..G..A.K.LQS.E.H.ED.S......T..CS...K.E...V...S..MLERNG..K......K.P...MGSPRE..RMMEA...TS-..V...N...SSV...AS..V. PRDM7_pteVam ..K.A............G.......MKRS....S..G..A.K.LQS.E.H.ED.S...--.N..RS...K.E...V...S...LERN...K.F....K.P...MGSPREY.RMMEA...TS-..V...N...SSV...AS..VI PRDM7_myoLuc ..K..........V.....T.....GKR....E...GAPAGN.LQSEE.G.ER.......QTG.HG...K.E...V.G.S...L.R....GT...SFK.PNRHMGSSSER.R.RE....T.-.NV.HKN.....V..KRSKSVT PRDM9a_bosTau .SK.K....A...............VQ......T.L.P.A.DYLQ.E.....S.....R-Y...HSPS.KPE.R.V.D.PQ..L....LK.....S.YSPR..MGASGVH.R.TE.-..TS-..P.........M.A.VSG..K PRDM9b_bosTau .SK.K....A...............VQR.....T.L.P.A.D.LQ.E.....N.....R-Y...HSPS.KPE.RKA.D.PQ..L...KLK.....S.YSPR..VGRSGVH.R.TE.-..TS-............M.A.VSG..K PRDM9d_bosTau ..K.K.Y..A..C.S..........VQR.......L.P.IGD.LQ.E.....S.....R-Y...HSLS.KPE.R.P...PH..L.G..PK...T.S.Y.P...MGGSEVH.RMTE.-..TS-............MEA.VSG.V. PRDM9e_bosTau ..K.K.Y..A..C.S..........VQR.......L.P.IGD.LQ.E.....S..E..R-Y...HSLS.KPE.R.P...PH..L.G..LK...T.S.Y.P...MGGSEVH.RMTE.-..TS-............MEA.VSG.V. PRDM9a_oviAri ..K.K....A....S..........VQRS......L.P.P.D.LQ.E.....K.....R-Y...HSPS.KPE...P...PH..L.G..LK...T.S.YTP...MGGSEVH.KMTE.-..TS-......N.....MEA.VSG.V. PRDM9b_oviAri .LK.K....A...P..........YVQP.......L.P.A.D.LQ.E.....N..E..--Y...HSPS.KPE.CKA...PPW.L..MSV-...M.S.YSP...MRGSETHYRMTE.-..TS-.......I....M.T.VSG..K PRDM9d_munMun ..K.K....A....T..........IQCS..P.T.L.P.E.DLLQ.E.....N.....R-Y...HSPS.KPE.H.A.D.PQ..L....LK.....S.CSPR..MGGSGVH.RMTE.-..TS-.....G...T.LT.A.VSG.MK PRDM9c_munMun ..K.K....A...............IQRS....T.L.P.E.DLLQ.E.....N...--R-F...H.PS.---------.PQ..L....LK.....S.YSPR..MGGSGVH.LMTE.-..TS-H........T.LM.A.VSG.M. PRDM9b_munMun ..K.K....A...............IQRS....T.L.P.E.DLLQ.E.....S...--R-Y...HSPS.KPE...A.D.PQQ.L....LK.....S.YSPG..MGGSGVH.RMTE.-..TS-.........T.LT.A.VSG.M. PRDM9a_munMun ..K.K....A......T........IQRS..A.T.L.P.E.NLLQ.EH....S...--R-Y...HSLS.KPE...A.D.PQ..L....LK.....S.YSPG..MGGSGVH.RMKD.-..TS-.........T.LT.A.VSG.M. PRDM9a_odoVir ..K.K....A...............IQCS....TP..P.E.DLLQ.E.....N...R---Y...HSPS.KPE...A.D.PQ..L....LK.....S.YSPG..MGGSGVH.--------------------------------- PRDM7_turTru D.K.K.Q..G..........I....V.CS....V...T.A.DRVQ.E.....Y..R...-Y...HS.SNKPEC..V...S...L.R..LG.......SSP...MGSSRAH.RMMEAG..T.-..V...A....LI.A.VS.VVK PRDM7_susScr ..K.K..............R.....V.RS....S...A.A.RGLQ.EG...DN.Q...P-YP..HS.DGTSES.DV..GS..FLERR.L.KT...S.YAPE..MRSSRVR.RMTE......-..V......T..TVA.ES---- PRDM7_lamPac --E.K.YL.................VK..........TAAGR.LE.E.....N..E...-...QHS...KPE...A...S..FL.R..L....G...YSH...MGNSRVHDRMIE....T.-..V..K......TWA.VS.TVE PRDM7_sorAra ..K...Y...C......N.....R.V..S...L...GT.A.T.PKSVNF...D...W..HSDPDEP...KLENHKS.G.S.....RMG.K...T..PNLRSSKMGSSNKH.T.MDKINTG--..E..K..YRV.A.I..P.... PRDM7_echEur ..K..............A....N..VK.S.......GT.T.KQPQVEN..LSN....K.-..NF.NQH.STES..AI.K....L.M.K.KT..NG..KLP.E.IGSSREH.KTKE..-.NSC..M.....SE.LV.L..S..V- PRDM9_homSap ..K.........C............V.R..S..NF.GP.A.KLLQ.EN....D...E..-YP..HSR..KT....I...S.L.N.RTW..E......S.P...MGSCRVGKR.ME..SRT.-..V..GN.....V...IS..AK
PRDM7_canFam VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECGrGFIQRSNLSIHQRTHTGEKP PRDM7_canLup VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECGrGFIQRSNLSIHQRTHTGEKP PRDM7_canAur VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECGRGFTQRSTLNEHQRTHTEEKP PRDM7_lycPic VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRGFTHRTNLIIHQRTHTGEKP YVCRECGrGFIQRSNLSIHQRTHTGEKP PRDM7_canMes VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRDFTHRTNLIIHQRTHTGEKP YVCRECGrGFIQRSNLSIHQRTHTGEKP PRDM7_speVen VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECgRGFTHRTNLIIHQTTHTGEKP YVCRECGrGFIQRSNLSIHQRTHTGEKP PRDM7_vulVul VKYRGCGRGFNDRSHLSRHQRTHMGENP YVCRECGRGFTHRTNLIIHQRTHTGEKP YVSWECGRSFTRRSNLITHQRTHTGEKP PRDM7_neoVis VKYRGSGQGFDDRSHLSRHQRTHKEEKP SVGKELRREFIHKSVLVTHQRTHTEALP PRDM7_musPut VKYRGSGQGFDDRSHLSRHQRTHKEEKP SVGKEPRREFIHKSVLVTHQRTHTGEKP YVCRECGRGFTQRSHLIRHQR PRDM7_ailMel VKYRGCGRDFSDRSHQSGHQRRH-QKKP SVCKKVKREFSHKSVLITHQRTHTGEKP YVCRECGRGFTQRSNLIRHQRTHTGEKP PRDM7_felCat IKNRGCEQGFNDRSHFSRHQRTHKEEKP SVCNEFRRDFSHKSALITHQRTHTGEKP YVCRECGRGFTQRSNLFRHQRTHTGEKP PRDM7_equCab VQYGGCGRGFNDRASLIKHQRTHTGEKP YVCRECEQGFTQKSSLIAHQRTHTGEKP YVCRECEQGFSEKSHLIRHQRTHTGEKP PRDM7_pteVam VKYGGCEHGFDDGSHLIMHQRTHSGEKP FVCRECERGFSKKSNLITHQRTHSGEKP FVCRECERGFTRKSSLITHQRTHSGEKP PRDM9_pteVam VKYGGCGHGFDDGSHFIRHQRTHSGEKP FVCRECERGFNEKSSLTMHQRTHSGEKP FVCRECE-GFSVKSSLIRHQRTYSGEKP PRDM7_myoLuc IKHGGCGQGFNDGSHIDTHQRTHSGEKP YICRECGGFTHKSDL IRHQRTHSQENP YVCRECGRGFRDRSTLITHQRTHSGEKP
gene_genSpp %id chr strand start stop span PRDM7_canFam 82% chr5 ++ 66560684 66567275 6592 dog CAD1 75% chr5 ++ 66571832 66581008 9177 GAS8 93% chr5 +- 66587321 66604940 17620 PRDM7_ailMel 100% GL193502 +- 628987 644235 15249 panda CAD1 73% GL193502 +- 620344 624223 3880 GAS8 91% GL193502 ++ 594843 609901 15059 PRDM7_felCat 100% Un_ACBE01450414 +- 10493 13105 2613 cat CAD1 75% Un_ACBE01450414 +- 3902 4280 379 GAS8 PRDM7_equCab 100% chr3 +- 36378853 36387224 8372 horse GAS8 93% chr3 ++ 36348528 36361906 13379 PRDM7_pteVam 100% ABRP01250178 +- bat GAS8 ABRP01250178 ++ PRDM7_myoLuc 100% AAPE02062260 +- bat GAS8 AAPE02062260 ++
Pecoran ruminants (cow, sheep, muntjak) present a vastly more complicated situation. Cows -- even in the revised assembly -- have a PRDM7 pseudogene adjacent to GAS8 accompanied by 5 PRDM9 copies in other locations (all distinct from the primate cadherin secondary site). This is neither a recent development nor an artifact of domestication because a similar expansion is seen in provisional assemblies of sheep and muntjak (wild deer) but not dolphin, pig or vicuna, dating the expansion to stem pecoran ruminant within artiodactyls. It is not clear which if any (or several acting in tandem) of these gene copies play a role in recombination -- the primate paradigm for meiotic markup is not immediately applicable to these species.
Atlantogenata (Afrotheres + Xenarthra) have yet another history. Elephant (best of the five available assemblies) has three loci: an old PRDM7 pseudogene in GAS8 syntenic position, a seemingly functional PRDM9a with 12 terminal zinc fingers and novel syntenic location, and a fairly recent pseudogene PRDM9b. Extinct mammoth shows the same three genes with the same pseudogenization pattern. Although the sequences diverged separately after speciation, three identical inactivating mutations occur in both mammoth and elephant but not hyrax, thus dating gene loss within afrothere speciation. This is shown for exon 9 below:
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGR 1 PRDM9_conSeq wildtype consensus reference 1 YVNCIQD*KEQNLVAFQYHRQIFHWTCCTIRPGCELLVWYGDNYSQELGIKWGSR*KKELTSGT 1 PRDM9b_loxAfr gg bad acceptor, early stop codon, internal stop codon 1 YVNCTRDKEEQNLVAFQYHRQIFYWTCHTIQPGCelLVWYGDNYGQELGIKWGSR*KKELTSGT 1 PRDM9b_mamPri gg bad acceptor, two 1 bp deletions, internal stop codon 1 YVRRARDTEERNLVAFQYHRQIFYRTCCTVRPGCELLVWRGAEDSQALG SRRTMELTSQK 1 PRDM9b_proCap pseudogene with 4aa deletion 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRTIQPDCELLVWYGDEYGQELGIKWGSRWKKELTSGT 1 PRDM9a_loxAfr wildtype 1 YVNCARDEEEQNLVAFQYHRQIFYRT 1 PRDM9a_mamPri fragmentary coverage 1 YVNCARDEDEQNLVAFQYHGQIFYRTCRPVQPGCELLVWYGDEYGQELGIQRGSRQMKALSSQT 1 PRDM9a_proCap 17 zinc fingers 1 YVNGTQDEKEQNLVFFQYHRQIFYQTCYAVWPGCQLLVWYRDECGQELGIKWDNRGKKEFTVGT 1 PRDM7_loxAfr bad acceptor, bad donor 1 YVNGTQDEKEQNLVFFQYHRQIFYQTCYAVWPGCQLLVWYRDECGQELGIKWDNRGKKEFTVGT 1 PRDM7_mamPri bad acceptor, bad donor, 1 synon bp difference 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRAIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAEK 1 PRDM7_choHof wildtype 1 YVNCAWDDKEQNLVAFQYHRQIFYRTCRTIRPGCELLVWYGDEYGQELGIKWGSKWKKEFMTGT 1 PRDM7_dasNov wildtype
Correct gene tree for PRDM7 and its spin-off PRDM9s
At left is the gene tree summarizing the evolutionary relationships of the 80-odd PRDM7/PRDM9 homologs currently available from GenBank genome and targeted sequencing projects. Since the mammalian species tree is well-known, the gene tree can be clamped to it rather than derived ab initio (which won't work because some loci that must be included are pseudogenes in various states of degeneration.
The gene tree shows PRDM7 (the fundamental gene here) spinning off various copies of itself at various times in various lineages over the last 102 million years of placental mammal evolution. Each spin-off has been offset and given a slightly different nomenclature to remind us that the primate PRDM9 gene duplication (called PRDM7L9 in the figure) should branch off at old world monkey and bear no special relationship to unrelated spin-offs in artiodactyls and afrotheres. The genes without offsets are conventional orthologs in the GAS8 PRDM7 qTer syntenic position, which has been stable for several billion years of summed branch length geologic time.
These spin-offs had different subsequent histories. In some lineages, like primates, the spin-off PRDM9 took over from a pseudogenized parental gene PRDM7; in others, it became a pseudogene; in others, both genes seem to have persisted.
Note neither carnivores nor rodents ever had a gene duplication of PRDM7. These species do not now contain a counterpart to human PRDM9 and never did. There is no evidence whatsoever for surviving pseudogene debris between CDH12-CDH10 (the syntenic location of all primate PRDM9), despite the extreme sensitivity possible with localized tBlastn searching.
The fact that neither dog nor mouse has PRDM9 is not about naming conventions. It is about shifting functionality and shifting mechanisms for initiating meiotic recombination over evolutionary time. To discuss that intelligently, the genetic loci involved are best given names that are in themselves informative about gene evolutionary relationships, ie be compatible with their placement in the gene tree. That in a nutshell it is wrong to call the dog homolog PRDM9. In fact, it clusters by its synteny with the PRDM7s (including human) whereas the primate PRDM9s are off on their own subtree. (Put another way, if this is dog PRDM9, then where is dog PRDM7?)
Human gene names are set by definition by HUGO and used exclusively in scientific journals by international agreement, the idea being to create a stable terminology (human genome is complete) and to avoid endless lab-specific synonyms, use of greek letters, roman numerals, subscripts, superscripts, upper/lower case, hyphens etc unsuitable for the bioinformatic era.
HUGO attempts to give full length paralogs (above a certain percent identity) the same base name followed by consecutive numbering. This is not done consistenly as seen from PRDM* which is a pseudo-family of 16 genes with only a small PR(SET) domain in common (or PRNP/PRND which as full length homologs should have been PRNP1 and PRNP2). Pseudogenes are sporadically named. Despite the many flaws in HUGO names, it is unlikely that they will ever be significantly revised from where they are now.
However the international agreement does not provide for official names or acronyms for the corresponding proteins and here the Wild West usage still prevails in journals. Just attaching a p as in PRDM9p to the HUGO name would work but people who work only at the protein level don't cooperate. However there is increasing pressure to standardize to allow computer mining of the biomedical literature. PubMed could require abstracts to contian a tag that would allow Blast searches against them (ie get around nomenclature variation). So far that hasn't happened.
While human gene names are by definition correct, their underlying gene models are not. HUGO does not involve itself in gene curation but simply defers to a RefSeq at NCBI (which has no formal mechanism to correct errors). There are legitimate issues involving the significance, if any, of alternative splices and exon skipping and when something is a pseuogene. Thus human olfactory genes are difficult to sort out; in folate metabolism, a former DHRF pseudogene got upgraded to DHFL1 only in Oct 2011.
All of the above just applies to humans, no other vertebrates. Mouse has its own official gene nomenclature committee (based at Jackson Laboratories). Here lower case is used for orthologs of HUGO gene names when possible (eg Prdm7) which doesn't scale to the other 5400 mammals.
So how should genes be named in other vertebrates? Note humans have lost quite a few genes relative to other species (120 or more relative to last common ancestor with chicken, including whole subfamilies of opsins). So those genes cannot inherit names from HUGO human nomenclature unless it is ghosted forward to include lost genes.
However in other mammals, simple 1:1 syntenic orthologs to human genes can be given the same name (eg dog or cat or panda PRDM7). This may covers 15,000 genes, a good start. For a segmental translocative duplication to a different chromosome like the PRDM7 -> PRDM9 case, PRDM7 is the parent gene and human PRDM9 the secondary derived feature. The syntentic gene in old world monkeys can safely be called PRDM9 as well. PRDM9 never existed in Carnivora and was not lost. Use of PRDM9 there would require a strange and different type of ghosted nomenclature than for lost genes.
So what to call the extra copies of PRDM7 in elephant? Recall they have an old PRDM7 pseudogene in GAS8 syntenic position, a fairly recent retroprocessed pseudogene, and a seemingly functional copy with 12 fingers which has nothing to do with the human PRDM9 syntenic location.
The name for this lineage-specific duplication in afrotheres should not confuse it with the unrelated lineage-specific duplication in primates (PRDM9) because these gene clades have no special relationship. Primate PRDM9 will branch out from the PRDM7 gene tree at primates, but the elephant copy from within afrothere PRDM7s .
The nomenclature proposal here follows the HUGO template for the folate gene DHFR. That is, PRDM7 and numbered PRDM7L for Lineage-specific duplication. Here the primate PRDM9 would be PRDM7L9, elephant might be PRDM7L1, PRDM7L2, PRDM7L3, PRDM7L4 etc etc for the duplications in artiodactyls.
The tree will need periodic revision as new mammalian genomes come in or as existing loci are re-interpreted. The Newick format that generates the tree is:
(((((((((((((PRDM7_homSap,PRDM7_panTro),PRDM7_gorGor),PRDM7_ponAbe),PRDM7_nomLeu),((PRDM7_macMul,PRDM7_macFas),PRDM7_papHam)),(((((._._.PRDM7L9_homSap,._._.PRDM7L9_panTro),._._.PRDM7L9_gorGor),._._.PRDM7L9_ponAbe),._._.PRDM7L9_nomLeu),((._._.PRDM7L9_macMul,._._.PRDM7L9_macFas),._._.PRDM7L9_papHam))),(PRDM7_calJac,PRDM7_saiBol)),PRDM7_tarSyr),(PRDM7_micMur,(PRDM7_otoGar,._._.PRDM7L8_otoGar))),PRDM7_tupBel),((((((PRDM7_musMus,PRDM7_ratNor),PRDM7_musMol),PRDM7_criGri),PRDM7_dipOrd),PRDM7_speTri),(PRDM7_oryCun,PRDM7_ochPri))),((((((((((((PRDM7_canFam,PRDM7_canLup),PRDM7_canAur),PRDM7_lycPic),PRDM7_canMes),PRDM7_speVen),PRDM7_vulVul),((PRDM7_neoVis,PRDM7_musPut),PRDM7_ailMel)),PRDM7_felCat),PRDM7_equCab),(PRDM7_myoLuc,(PRDM7_pteVam,._._.PRDM7L7_pteVam))),(((((((PRDM7_bosTau,._._.PRDM7L5_bosTau),PRDM7_oviAri),(PRDM7_munMun,PRDM7_odoVir)),((((._._.PRDM7L1_bosTau,._._.PRDM7L1_oviAri),._._.PRDM7L1_munMun),((._._.PRDM7L2_bosTau,._._.PRDM7L2_oviAri),._._.PRDM7L2_munMun)),(((._._.PRDM7L3_bosTau,._._.PRDM7L3_oviAri),._._.PRDM7L3_munMun),(._._.PRDM7L4_bosTau,._._.PRDM7L4_oviAri)))),PRDM7_turTru),PRDM7_susScr),PRDM7_lamPac)),(PRDM7_sorAra,PRDM7_echEur)))
,(((((((PRDM7_loxAfr,._._.PRDM7L2_loxAfr),PRDM7_proCap),(._._.PRDM7L1_loxAfr,._._.PRDM7L1_proCap)),PRDM7_echTel),(PRDM7_dasNov,PRDM7_choHof)),((PRDM7_macEug,PRDM7_monDom),PRDM7_sarHar)),PRDM7_ornAna));
The genusSpecies acronyms are in alphabetic order below: ailMel Ailuropoda melanoleuca (panda) bosTau Bos taurus (cattle) calJac Callithrix jacchus (marmoset) canAur Canis aureus (golden jackal) canFam Canis familiaris (dog) canLup Canis lupus (gray wolf) canMes Canis mesomelas (black-backed jackal) choHof Choloepus hoffmanni (sloth) criGri Cricetulus griseus (hamster) dasNov Dasypus novemcinctus (armadillo) dipOrd Dipodomys ordii (kangaroo rat) echEur Erinaceus europaeus (hedgehog) echTel Echinops telfairi (tenrec) equCab Equus caballus (horse) felCat Felis catus (cat) gorGor Gorilla gorilla (gorilla) homSap Homo sapiens (human) lamPac Lama pacos (llama) lycPic Lycaon pictus (painted dog) macEug Macropus eugenii (wallaby) macFas Macaca fascicularis (crab-eating macFas Macaca fascicularis (crab-eating macMul Macaca mulatta (rhesus) micMur Microcebus murinus (lemur) monDom Monodelphis domestica (opossum) munMun Muntiacus muntjak (muntjac) musMol Mus molossinus (wild mouse) musMus Mus musculus (mouse) musPut Mustela putorius (ferret) myoLuc Myotis lucifugus (bat) neoVis Neovison vison (mink) nomLeu Nomascus leucogenys (gibbon) ochPri Ochotona princeps (pika) odoVir Odocoileus virginianus (deer) ornAna Ornithorhynchus anatinus (platypus) oryCun Oryctolagus cuniculus (rabbit) otoGar Otolemur garnettii (galago) oviAri Ovis aries (sheep) panTro Pan troglodytes (chimp) papHam Papio hamadryas (baboon) ponAbe Pongo abelii (Sumatran pteVam Pteropus vampyrus (bat) ratNor Rattus norvegicus (rat) saiBol Saimiri boliviensis (squirrel monkey) sarHar Sarcophilus harrisii (tasmanian devil) sorAra Sorex araneus (shrew) speTri Spermophilus tridecemlineatus (squirrel) speVen Speothos venaticus (bush dog) susScr Sus scrofa (pig) tarSyr Tarsius syrichta (tarsier) tupBel Tupaia belangeri (tree shrew) turTru Tursiops truncatus (dolphin) vulVul Vulpes vulpes (red fox)
Marsupials and platypus: the mystery of exon 5
Tracking PRDM7 back to marsupials and beyond is problematic. The three available marsupial assemblies are seriously incomplete, causing gene prediction issues as exons are spottily represented and spread over multiple small contigs which cannot be tiled up into full-length genes, much less yield syntenic information. Because PRDM7/9 contain domains found in many other chimeric proteins, isolated exons cannot always be assigned correctly to their parent gene.
Further, some exons in PRDM7/9 have weak amino acid conservation and so fail to give definitive blast matches to placental queries, a problem exacerbated for short exons and decayed pseudogenes (opossum). No expression data exist to bridge uncertain regions, meaning missing diverged exons cannot be located. Because the domains here occur widely in other combinations in other proteins, a full length marsupial sequence is critical to testing whether the domain shuffle resulting in PRDM7 and PRDM9 was a placental innovation.
The most favorable situation occurs in the Monodelphis domestica assembly. Although exons 1 and 5 are missing, eight of the ten expected exons are readily located in a single assembly region of length 33,449 bp containing a single internal gap (estimated at 270 bp). It is not surprising that exon 1 cannot be located because it contains no known Pfam domain or reason for fixed length and diverges rapidly in placentals. However locating exon 5 is important for distinguishing between two adjacent small genes evolving into a single fused gene only in the placental branch versus a full length gene already present in the last common ancestor.
Unless exon 5 lies within the assembly gap, it should be locatable in the 25,548 bp separating exon 4 and exon 6 (of which 8,263 bp remains after application of RepeatMasker). However blastx against a panel of 54 exon 5 sequences from placental mammals fails to give any suggestion of match in any species, despite plausibly adequate length (all placental exon 5 sequences have 52 amino acids).
Gene prediction tools such as GenScan, NScan, Ensembl and Gnomon give unsatisfactory results: a few exons are correctly predicted but are otherwise embedded in time-wasting rubbish. The poor reliability of these tools does not justify GenBank clutter (eg XM_001369137) providing their predictions. The 46-species whole genome alignment at UCSC (starting with PRDM7/9 'ProteinFasta' link at the description page) is a better starting point.
Here it should be noted that exon 5 has not diverged especially rapidly from the last common ancestor of placentals. Aligned to human, the full range of sequences has overall identity of 69%. Exon 5 has a number of invariant and semi-invariant residues, only possible over this time span if maintained by selective pressure. Thus it has some function even though it contains no known Pfam domains and has no crystallographic structure match.
Because exon 4 has a splice donor of phase 0 and exon 6 a splice acceptor of phase 2, the putative exon 5 in marsupials must take the form 0 xxx...xxx 1 to conserve reading frame. This rules out non-use of exon 5 in marsupials (alternative splicing) followed by mutational decay to unrecognizability. In the scenario of two adjacent genes not yet fused, the distal region would need a new exon containing the initial methionine and phase 1 splice donor because no iMet occurs in the extended reading frame of exon 6.
The opossum gene is peculiar in that 7 of the 8 exons available are quite conventional in sequence but the terminal zinc finger exon is completely broken up by frameshifts and stop codons and barely recognizable. It could not be used to initiate meiotic recombination, yet no substitute homolog is at hand. The other exons return only PRDM7/9 as significant matches when back-blasted against the human genome establishing that they have not been confused with the many hundreds of partial homologs with KRAB, SSXRD, PR (SET) or C2H2 domains. However human (and other placentals) could easily have lost even better blast matches since divergence from marsupials. Thus it remains unclear whether marsupials have a full length counterpart to placental PRDM7.
The Sarcophilus harrisii assembly is missing the same two exons but has a conventional terminal exon with an intact zinc finger region of seven repeats (with two distal frameshifts however). Here exons 2 occurs in contig AFEY01202902 and exons 3-4 in AFEY01156721 with 1,436 bp left over to host exon 5; exons 6-10 are found in a third contig AFEY01386448 with 8,331 bp available upstream for exon 5. It can't be established that these contigs are actually adjacent in the genome. The six exons comparable between tasmanian devil and opossum are 82% identical to each other as proteins and 67% identical to those of human, not indicative of anomalous or especially rapid evolution in the context of entire proteome rates.
The Macropus eugenii (wallaby) assembly is least complete, with no contig containing more than a single exon. Here exons 1, 4, 5 and 8 are missing altogether but the terminal zinc finger exon is intact with 7 C2H2 domains. It is worth noting that the exon 10 is so long and distinctive with its phase 2 reading frame and early zinc finger that there is no possibility of confusing it with the closest human homologs (HKR1, ZNF133, ZNF169, ZNF343, ZNF589). However, humans could have lost an even better homolog of this exon.
The gene adjacent to PRDM7 in mammals, GAS8, is an ideal probe, being single-copy and quite conserved in vertebrates. In the ancestral placental mammal, GAS8 and PRDM7 are convergently transcribed. Thus a marsupial contig containing the last exons of GAS8 might contain the last exon (or 3' UTR) of PRDM7. Even a partial exon or pseudogene remnant could be recognized with great sensitivity in such a contig. However none of the marsupial GAS8 contigs contain any information on PRDM7 or any other gene.
The situation in platypus and echidna is curious according to 18 recent articles. Note first that the chain of ten X and Y chromosomes (which is unprecedented in mammals) segregates during meiosis into either an X chain or Y chain, requiring crossovers in the 9 paired pseudoautosomal regions. Homology of key genes is to chicken, not theran mammals whose sex chromosomes thus arose after divergence at 165 million years. As with meiosis initiation, sex chromosomes seem never to stop evolving.
It is not clear whether some form of PRDM7/9 is operative outside of placental mammal -- meiotic events have not been experimentally characterized to date in either marsupials or monotremes. Bird, alligator, and lizard (7 genomes) all lack candidate orthologs. Thus it is uncertain whether the massive restructuring of sex determination around this time correlates with the switchover to PRDM7/9 for meiotic recombination (in view of the sex chromosome recombination bottleneck) or is simply coincidental.
In terms of platypus PRDM7/9 candidate orthologs, only distal exons 6-10 can be reliably recognized in the current assembly, ie KRAB, SSXRD and exon 5 are missing but the knuckle, PR and zinc finger domains are present with 3-4 repeat units. However the early zinc finger in the last exon is not present. Nonetheless, the best back-blast to human is still PRDM7/9. These exons occur in two tandem copies on the same strand but differ significantly from each other and so do not represent mis-assembly duplications. The intervening area is gapless so the missing exons should be locatable if present.
However they are not. Upon blastx of the repeatmasked sequence against Genbank tetrapod sequences, no matches occur, other than three worthless platypus gene models (XP_001507240, XP_001509482, XP_001509433) that predict earlier exons which however are wholly lacking in any support in any other species. Thus it appears that the gapless region does not contain any counterpart to exons 1-5 of theran mammals. Either this region has been lost in platypus or it is a stand-alone shorter distal version of PRDM7/9.
The first identifiable exons begins with the expected phase 2 reading frame in both tandem copies and do not contain an in-frame methionine upstream prior to a stop codon. Hence there must be at least one earlier exon. However tblastx of the appropriate regions of repeatmasked marsupial and platypus again does not identify noteworthy candidates.
Perhaps the corresponding ancestral region was shuffled together with a gene providing the proximal regions in the theran branch only, giving rise to the full length gene there. However tblastn queries of the platypus assembly, while locating numerous appropriate KRAB_A domains with the correct 0 xxx...xxx 1 reading frame that back-blast to other human proteins, do not find counterparts of the exon 1-5 region beyond exon 2. Hence there is no obvious donor for the proximal half of PRDM7/9.
Given that the PRDM and zinc finger families are greatly expanded with extensive domain shuffling in mammals with difficulties already tracing back PRDM7/9 to marsupials and monotremes, it comes as no surprise that bird, lizard and frog genomes shed no further light on the evolution of this gene. The situation in non-placental mammals could theoretically be resolved by sequencing transcripts, but these are exceedingly rare for PRDM7/9 even in placentals and so will not emerge unless explicitly sought.
Conservation of exon 5 within placentals; invariant residues in red PRDM9_homSap GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL PRDM9_panTro .......N.........GMP....T............P.............. PRDM9_gorGor .....................................P.............. PRDM9_ponAbe .......N.........G.Q....T............P..........T..I PRDM9_nomLeu .................GA..................P.............. PRDM9_macMul .......N.......V.GM.....T............P...R.......... PRDM9_papHam E...T............G.P...ST.........A..P.............. PRDM7_calJac .......G......K..G...V..T..P.........P.............. PRDM7_micMur ...R.PL.DG.......G......T.....P......PR..........R.. PRDM7_otoGar ...R.PL.DG.......GP.S.P.I.....H..HM.SPR.........GR.S PRDM7_tarSyr ...R.PL.IV.......EM.....T.D....W......R.....E....K.. PRDM7_oryCun ...RLPVN.........GI.....TT...ED...SF.PK.TR......TR.. PRDM7_ratNor ET.RMPL.DK..V..VFGIE....T....H.....CSPE.GN.....FGK.. PRDM7_musMus ESSRMP..G..NV..G.GIE....T....HV.....SLE.GN......GK.. PRDM7_speTri LK.EVLL..........G......T.....V......LR...A.R....R.. PRDM9e_bosTau ..SR.PL.K.......PGA.K..KT..CK....L.P.PRK.R.PE..P.Q.V PRDM9c_oviAri ..S..LV..K.....MPGASK..KTR.PK...I..PAPR.P...E..P.Q.V PRDM9a_munMun ..SR.PLIK.......LGA.K.MKT...K...N..PHPRK.R.P...P.Q.V PRDM7_turTru AV.PVPL.......K.PGA.Q.QK...PA...S.AP.P.A....AW.T.Q.. PRDM7_lamPac ...RGPL..Q.......G..KP.KT...G.....FP.L.......R...Q.. PRDM7_susScr SDSRVPL..K......LT..EVPET.......E....P......RRR.GQE. PRDM7_canFam .I.RVPL..K.......E..K...T.SP..G..S..LP.K.....H.T.Q.. PRDM7_felCat .THRVPL.K.....DF.E..K...T.....G.....LP.......H...R.. PRDM7_ailMel .I.R.PLR.........E..K...T....LG.....LP.......HD.LQ.. PRDM7_musPut .V.R.PL..........E..K...T....HD.....HP.......H..LR.. PRDM7_pteVam A..RVPL...P......VI....K......D....F.P.K..A.R....Q.. PRDM7_myoLuc AKSR.PL..........G.....TT.....T..T.P.P.........P.S.. PRDM7_equCab R.RT.PL....R.....G..K..KT.S...V......L....S.E....R.. PRDM7_sorAra .RSRTPI.....S....G.RT...TKCTK.....LF.P.......HY.KP.. PRDM9a_loxAfr .T...LLG.......V.G..I...TT..........SP......D.P..W.. PRDM7_echTel ...GV.LR...N..V..G..I..T.AEP..PH-.G..P...T..HE.L.Q.V PRDM7a_proCap .T...LLG.......V.G..I...TT..........SP......D.P..W.. Consensus GMPRAPLSNESSLKELSGTANLLNTSGSEQAQKPVSPPGEASTSGQHSRQKL
Comparative genomics: sequence availability
As of Sept 2011, some 72 PRDM7 and PRDM9 genes from 45 species can be recovered from mammal genome projects. The encoded proteins are parsed into exons in the Curated reference sequences section. Due to gaps in coverage, full length gene models could not always be established.
There has been no gain or loss of introns -- all genes have the same identically phased ten exons. No retroprocessed (intronless) genes occur in any species despite transcription in germline tissues. However 83 of 710 expected exons could not be found for lack of coverage or (for marsupials and monotremes) possibly too much divergence.
In low coverage genomes, internal frameshifts and stop codons cannot easily be distinguished from sequence error even by consulting raw trace reads. However multiple disabling changes accompanied by an amino acid substitution pattern incongruent with conservation profile are strong evidence of pseudogenization, at least past the point of the first inactivating mutation. In chimeric proteins, the proximal region of the protein could continue to be functional.
Since typically only one individual of unspecified gender of a species is sequenced on just one of two relevant autosomal chromosomes, an aberrant gene could reflect a bad heterozygous allele, an atypical homozygous individual (who might have impaired meiosis), or a balanced polymorphism that advantageously reduces copy number. A population survey is necessary to distinguish between these possibilities and overall SNP variation. In some genes, the zinc finger array seems too short to have sufficient site specificity. However these are known to contract and expand in the two intensively studied species (mouse, human), so here too the sequence from a single individual can be misleading. Without this data, it can difficult to say whether a given PRDM7/9 locus is a pseudogene.
Supporting transcripts do not resolve the issue, first because pseudogene can continue being transcribed for millions of years after losing all functionality at the protein level and second because PRDM7 and PRDM9 are barely represented among the millions of mammalian transcripts at GenBank. That rarity might be explained by low levels of transcription in tissue types not widely used as experimental sources. However testis is frequently studied and one or more members of this gene family is essential for meiosis. This illustrates the futility of undirected transcript sequencing projects for determining the full coding potential of the genome. Global expression chips to date have not produced results here either.
The transcripts from mouse, rat and pig do not support the widely propagated concept that PRDM7/9 function solely in meiosis (which would limit them in effect to testis) as most transcripts arise elsewhere. In mouse, the PRDM7 role in meiosis has strong experimental support, yet many transcripts come from non-meiotic tissues. Human PRDM9 experimental transcripts mostly derive from a single unpublished 2011 project entitled "Exhaustive RT-PCR and sequencing of all novel TWINSCAN predictions in human" which unhelpfully pooled tissue from adrenal gland, bone marrow, brain, cerebellum, brain (whole), fetal brain, fetal liver, heart, kidney, liver, lung, placenta, prostate, salivary gland, skeletal muscle, thymus, thyroid, trachea, uterus, and spinal cord with testis.
Transcripts at GenBank on 22 August 2011 (est database): DB452778 PRDM9 homSap testis DB636359 PRDM9 homSap testis DB024448 PRDM9 homSap testis DB080053 PRDM9 homSap testis DT932634 PRDM9 homSap pooled including testis DT932633 PRDM9 homSap pooled including testis DV080525 PRDM9 homSap pooled including testis DV080526 PRDM9 homSap pooled including testis DV080328 PRDM9 homSap pooled including testis DV080173 PRDM9 homSap pooled including testis DV080174 PRDM9 homSap pooled including testis DV080327 PRDM9 homSap pooled including testis BU194881 PRDM9 homSap melanotic melanoma AL704902 PRDM9 homSap not reported GU216230 PRDM7 musMus testis FJ212287 PRDM7 musMus testis HQ704390 PRDM7 musMus testis? HQ704391 PRDM7 musMus testis? CK032493 PRDM7 musMus placenta CJ235803 PRDM7 musMus amnion CN723438 PRDM7 musMus 4-cell embryo BI737497 PRDM7 musMus retina BB642583 PRDM7 musMus retina BC012016 PRDM7 musMus retina BC023014 PRDM7 musMus retina BG288443 PRDM7 musMus eye FM103467 PRDM7 ratNor body fat GO353654 PRDM7a bosTau 4-cell embryo EF432551 PRDM7 bosGru testis BX673635 PRDM7 susScr pooled including testis CO991452 PRDM7 susScr oviduct EW469934 PRDM7 susScr mucosal membrane
The table below shows the number of zinc fingers in the second column, phylogenetic clade in the third, and adjacent gene (synteny) in the fifth. The number and character of zinc fingers is quite variable in human populations and likely so in all mammals; the table provides that of the individual selected for reference genome project which may not be representative of the species.
These zinc finger arrays have been corrected in low coverage genomes for common sequencing errors -- frameshifts and premature stop codons arising from nucleotide run length mis-calls (eg, ggggg read as gggg) -- though they could actually represent valid mutant alleles in the heterozygous state (assuming the gene essential for meiosis). Indeed, these errors seem far more common than in what is seen in housekeeping genes for the same genome.
The PRDM7 genes are all orthologous in the classical sense (as can be seen by adjacency to the unrelated gene GAS8) but various PRDM9 genes arose as different lineage-specific segmental duplications so are orthologous within a delimited phylogenetic clade. There is currently no suitable nomenclature for different gene duplications in different clades of the same parental gene so they are just called PRDM9 here, with PRDM7 reserved to genes adjacent to GAS8. In some species such as mouse, chromosomal rearrangements have scattered syntenic relations and orthology remains slightly uncertain but the single gene in the genome probably represents simple descent from the single euarchontoglire PRDM7 gene.
- PRDM7: genes with ancestral location GAS8 synteny
- PRDM9: lineage-specific segmental duplications of PRDM7
- Pseudogenes: multiple disabling frameshifts and stop codons in parental gene (not a retrogene)
>PRDM9_homSap 13 prim gene CDH12 Homo sapiens (human) NM_020227 >PRDM9_panTro 19 prim gene CDH12 Pan troglodytes (chimp) GU166820 >PRDM9_gorGor - prim gene cdh12 Gorilla gorilla (gorilla) CABD02290264 >PRDM9_ponAbe 10 prim gene CDH12 Pongo abelii (orangutan) XR_093432 >PRDM9_nomLeu 10 prim gene cdh12 Nomascus leucogenys (gibbon) ADFV01015315 >PRDM9_macMul 9 prim gene CDH12 Macaca mulatta (rhesus) XM_001083675 >PRDM9_papHam 11 prim gene cdh12 Papio hamadryas (baboon) genome >PRDM7_homSap 3 prim gene GAS8+ Homo sapiens (human) genome >PRDM7_panTro 2 prim pseu GAS8+ Pan troglodytes (chimp) genome >PRDM7_gorGor 3 prim pseu GAS8+ Gorilla gorilla (gorilla) genome >PRDM7_ponAbe 4 prim gene GAS8+ Pongo abelii (orangutan) genome >PRDM7_nomLeu 5 prim pseu gas8+ Nomascus leucogenys (gibbon) ADFV01125891 >PRDM7_macMul 2 prim pseu GAS8+ Macaca mulatta (rhesus) genome >PRDM7_papHam 2 prim pseu gas8+ Papio hamadryas (baboon) genome >PRDM7_calJac 12 prim gene GAS8+ Callithrix jacchus (marmoset) XR_090591 >PRDM7_tarSyr - prim pseu gas8+ Tarsius syrichta (tarsier) ABRT011082008 >PRDM7_micMur 8 prim gene gas8+ Microcebus murinus (lemur) ABDC01433247 >PRDM7a_otoGar 10 prim gene GAS8+ Otolemur garnettii (galago) genome >PRDM7b_otoGar 8 prim gene GAS8+ Otolemur garnettii (galago) genome >PRDM7_tupBel 9 prim gene noDet Tupaia belangeri (tree_shrew) genome >PRDM7_oryCun 4 glir gene other Oryctolagus cuniculus (rabbit) genome >PRDM7_ochPri - glir gene noDet Ochotona princeps (pika) AAYZ01312269 >PRDM7_ratNor 10 glir gene PDCD2 Rattus norvegicus (rat) NM_001108903 >PRDM7_musMus 12 glir gene PDCD2 Mus musculus (mouse) NM_144809 >PRDM7_musMol 11 glir gene noDet Mus molossinus (wild_mouse) GU216230 >PRDM7_criGri 3 glir gene noDet Cricetulus griseus (hamster) AFTD01086355 >PRDM7_dipOrd - glir gene noDet Dipodomys ordii (kangaroo_rat) genome >PRDM7_speTri - glir gene noDet Spermophil tridecemlin (squirrel) AAQQ01308561 >PRDM9a_bosTau 7 laur gene noDet Bos taurus (cattle) NW_003053109 >PRDM9b_bosTau 5 laur gene noDet Bos taurus (cattle) DAAA02065087 >PRDM9c_bosTau - laur gene noDet Bos taurus (cattle) XM_002699750 >PRDM9d_bosTau 9 laur gene noDet Bos taurus (cattle) genome >PRDM9e_bosTau 9 laur gene noDet Bos taurus (cattle) genome >PRDM9e_oviAri - laur pseu noDet Ovis aries (sheep) genome >PRDM9d_oviAri - laur gene noDet Ovis aries (sheep) genome >PRDM9c_oviAri 4 laur pseu noDet Ovis aries (sheep) genome >PRDM9b_oviAri 2 laur pseu noDet Ovis aries (sheep) genome >PRDM9a_oviAri 9 laur gene noDet Ovis aries (sheep) genome >PRDM9d_munMun 4 laur gene noDet Muntiacus muntjak (muntjac) AC216498 >PRDM9c_munMun 15 laur gene noDet Muntiacus muntjak (muntjac) AC154919 >PRDM9b_munMun 13 laur gene noDet Muntiacus muntjak (muntjac) AC218859 >PRDM9a_munMun 7 laur gene noDet Muntiacus muntjak (muntjac) AC225653 >PRDM7_bosTau - laur pseu GAS8+ Bos taurus (cattle) genome >PRDM7_turTru 9 laur gene gas8+ Tursiops truncatus (dolphin) ABRN01441536 >PRDM7_lamPac 2 laur gene noDet Lama pacos (llama) scaffolds >PRDM7_susScr 9 laur gene GAS8+ Sus scrofa (pig) FP476134 >PRDM7_canFam 5 laur pseu GAS8+ Canis familiaris (dog) genome >PRDM7_felCat 11 laur gene GAS8+ Felis catus (cat) genome >PRDM7_ailMel 6 laur gene GAS8+ Ailuropoda melanoleuca (panda) GL193502 >PRDM7_musPut 3 laur gene noDet Mustela putorius (ferret) AEYP01035077 >PRDM7_neoVis 2 laur gene noDet Neovison vison (mink) JF288183 >PRDM9_pteVam 15 laur pseu noDet Pteropus vampyrus (bat) ABRP01232219 >PRDM7_pteVam 7 laur gene GAS8+ Pteropus vampyrus (bat) ABRP01250178 >PRDM7_myoLuc 6 laur gene gas8+ Myotis lucifugus (bat) AAPE02062260 >PRDM7_equCab 4 laur gene GAS8+ Equus caballus (horse) genome >PRDM7_sorAra 8 laur gene noDet Sorex araneus (shrew) AALT01000095 >PRDM9a_loxAfr 12 afro gene noDet Loxodonta africana (elephant) genome >PRDM9b_loxAfr 3 afro pseu noDet Loxodonta africana (elephant) genome >PRDM7_loxAfr 5 afro pseu GAS8+ Loxodonta africana (elephant) genome >PRDM7_echTel 5 afro pseu noDet Echinops telfairi (tenrec) genome >PRDM7a_proCap 17 afro pseu noDet Procavia capensis (hyrax) ABRQ01392668 >PRDM7b_proCap 13 afro pseu noDet Procavia capensis (hyrax) ABRQ01227339 >PRDM7_dasNov 9 xena pseu noDet Dasypus novemcinctus (armadillo) AAGV020462211 >PRDM7_choHof 2 xena pseu noDet Choloepus hoffmanni (sloth) ABVD01893961
Gene trees based on domains
PRDM9 is a chimeric protein consisting of 6 domains and linker regions. These domains occur in various combinations in many other human proteins without however known variability in domain order. The evolutionary relationships between all these proteins is necessarily complex, but taking the PR(SET) histone methylase as common denominator, the [gene tree at left emerges after structural alignment considerations.
While informative, this is really just a domain tree. A different tree would result based on the KRAB domain because it involves a different (though overlapping) set of proteins which had a partially independent history of duplication and shuffling from the PR(SET) domain. That precludes a meaningful joint tree based on KRAB + PR(SET) for those proteins that have both.The SSXRD domain has quite limited distribution but is considered further below. The knuckle and early zinc finger domains are rather short for domain tree inference, leaving presence/absence as the main consideration.
A domain tree based on the terminal zinc finger array is problematic due to long independent histories of expansion and contraction. Here the main handle is the C2H2 classification (based not only on residues binding zinc but also their spacing). Main other types of zinc fingers occur in the human proteome. Some blur into C2H2 but others -- like the intertwined CCHC and HCCC in DRMT1 -- are structurally quite distinct. DRMT1 is the sex-detemination gene in birds and a major regulator of gene expression in mammalian Sertoli and germ cells. It dramatically affects expression of mouse PRDM7 (called Prdm9) but apparently indirectly as the mouse gene lacks close-in upstream binding site according to genome browser wig tracks.
The traditional PR(SET) domain seems too small for an enzyme with such distinctive substrates so flanking sequence can be added consistent with observed amino acid conservation. Using S-adenosyl methionine as donor, PRDM9 places the third methyl group only on the fourth position lysine in mature histone H3 (which is actually position 5 prior to iMet removal: MARTKQTARK...), just one of this histone's 27 modified residues. There are many such epigenetic methylases in the human genome. PRDM9 has no applicable crystallographic structures, leaving undefined the residues involved in substrate binding and catalysis.
The histone orthology class, methylation position and methylation extent of these methylases correlates poorly with evolutionary grouping by PR(SET) domain (figure), suggesting gene duplications can readily diverge in their properties. PR(SET) domains can even lose catalytic competence yet retain recognition capacity and recruitment of other proteins. However loss of constraints might lead to anomalously fast divergence and so to misplacement in the domain tree.
The upper left corner shows variability in domain structure. While PRDM9 and PRDM7 share the same domains (an upstream KRAB domain is not shown), of PR-class homologs, PRDM11 shares only the PR(SET) domain despite nesting deep within the PRDM9 subtree. PRDM4 has both the PR(SET) and C2H2 domains, possibly sharing the early zinc finger in an exon beginning with a phase 2 splice acceptor (marked up with color in reference sequence collection). Overall however, PRDM9 and PRDM7 have no full length homologs with matching exon structure.
Note PR(SET) domain is even intronated differently within PR-class proteins, suggesting ancient divergence from a common pattern since intron gain/loss is exceedingly rare in vertebrates. These incongruities may have arisen from domain shuffling, gain and loss. Intron phasing provides a very important constraint on domain shuffling because the downstream reading frame must be preserved.
Intronation patterns of PRDM9 domains fit the standard eukaryote pattern: domains evolved first, introns inserted later at random sites. Domain shuffling might be even more pervasive if domains corresponded cleanly with exon breaks and all introns were phase 0. However this is almost never the case, another instance of what is sometimes called 'unintelligent design'.
The human PRDM9 sequence below is annotated in color for domains relative to exon breaks. The protein can be best understood in terms of concatenated domains, not all of which may be present in antecedent and descendant homologs. The first two domains KRAB and SSXRD likely interact with transcription factors, though these have not been specifically characterized in meiosis.
Each terminal zinc finger type C2H2 array potentially recognizes a specific trinucleotide and so a large concatenated array quite specific binding sites in the genome, though tolerance of nucleotide variability and overlapping interactions between adjacent units make it difficult to read out these sites precisely, despite immense efforts. However aberrant individual zinc fingers are common in arrays; not all contribute directly to dna binding specificity.
The concatenated C2H2 domains, conserved at the amino acid level so necessarily similar at the dna level, are apparently prone to replication slippage (or mis-registered gene conversion). Both processes can give rise to associated point mutations as well as leading to different repeat number distributions in human populations.
Taking the extremes of variations, it is a wonder that humans can still interbreed, yet Haldane's Rule so far has not been invoked as a cause of human infertility. PRDM9 allelic differences may provide an importent speciation barrier (ie, infertility of F1 males), yet introgression has been reported for denisovan, neanderthal and earlier African hominid dna into contemporary human genomes. By way of comparison, mice with 13 zinc finger repeats form sterile hybrids when crossed with mice differing merely by an extra repeat.
Many other unrelated proteins with internal repeats (such as the octapeptide region of the prion gene PRNP) are also affected by replication slippage. These events, though rare, have been intensively studied because they cause toxic gain of function (Creutzfeldt-Jacob disease). Repeat expansions here too are accompanied by localized point mutation. The PRNP repeats have anomalously high GC content prone to self-similarity loop-outs unlike the C2H2 repeats of PRDM9.
Both PRDM9 and PRDM7 contain a seldom-mentioned zinc finger early in the final exon, as annotated by SwissProt and readily found by the online domain tools such as SMART regardless of species. This domain conserves the four critical residues needed for zinc binding (and so the associated fold) but lacks the terminal cap TGEKP which otherwise serves to lock down a C2H2 zinc finger after it has scanned along genomic dna to an appropriate trinucleotide. The function of this early domain and the following 112 highly variable residues are unknown -- no demonstrably homologous sequence occurs in other human proteins with the possible exception of PRDM4 and PRDM15.
The main zinc finger array also resides in this long distinctive terminal exon of splicing phase 12 that has been shuffled together into various contexts during mammalian evolutionary time. For once, intron phase is not so informative because the preceding PR(SET) domain with its codon overhang of 1 bp can accept any shuffled domain with overhang of 2 bp and still maintain reading frame. Since the KRAB domain also terminates in a phase 12 splice site, proteins can also skip the PR(SET) domain entirely, as in ZNF133 and many others. Concepts such as paralogy and orthology thus need piecewise definitions in these composite proteins.
The first C2H2 of the main repeat region is proximally degenerate, beginning in VKY in all species (instead of YCE). The lysine cannot plausibly replace the usual cysteine for zinc binding though the other three needed residues are present and may suffice. This domain ends in a typical cap region TGEKP. Humans are the exception here where the conserved helix-ending proline has been replaced with leucine in the reference human genome, with unknown functional consequences. This replacement is not recent since it is found in all human populations including the extinct Denisovans (41 kyr) and the basal (70 kyr) bushman lineage for which fragment VKYGECGQGFSVKSDVITHQRTHTGEKL YVCRECGRGFSWKSHLLIHQRIH is available from read 20_@FQ2QD2002IAZ67.
Another overlooked zinc finger domain occurs in the same exon as the PR(SET), preceding it. Being short, it is sometimes called a zinc knuckle rather than finger. There can be no doubt about its occurence because a crystallographic study has confirmed the expected fold and zinc atom.
As noted, PRDM7 occurs immediately telomeric to the unrelated single-copy conserved gene GAS8 (with the two genes convergently transcribed). PRDM7 is otherwise the last gene on the q arm of its chromosome in many species which may predispose it to copy number dispersal events, which may in the past have resulted in juxtaposition and functional fusion to other genes. PRDM9 is not consistently present in placental mammals and each clade with it has a different syntentic location, suggesting numerous independent gene duplications (rather than many rearrangements and gene losses).
>PRDM9_homSap Homo sapiens (human) Q9NQV7 10 exons chr5:23,509,579 span 18,301 bp KRAB SSXRD zinc knuckle PR(SET) early ZNF C2H2 cap 0 MSPEKSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKL YVCRECGRGFSWKSHLLIHQRIHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRDKSHLLRHQRTHTGEKP YVCRECGRGFRDKSNLLSHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRNKSHLLRHQRTHTGEKP YVCRECGRGFSDRSSLCYHQRTHTGEKP YVCREDE* 0 -1 23 6 traditional numbering of dna recognizing amino acids LYCEMCQNFFIDSCAAHGPPTFVKDSAV alignment of zinc knuckle HPCPSCCLAFSSQKFLSQHVERNH alignment of pre-array zinc finger * * * * zinc liganding positions
Segmental duplications creating PRDM9s from PRDM7
In humans, PRDM9 and PRDM7 are related by a 26 kbp segmental duplication of chr16:90123419-90147718 that begins about 8 kbp upstream of the start codon and continues through most of the 3' UTR. Since the retroposon patterns are nearly identical, the duplication must be fairly recent. The overall percent identity of non-coding dna is about 93%, again inconsistent with either early (stem placental) or late divergence (great ape). The duplication contains a potentially diagnostic 1845 bp retroposon-free region upstream of the first coding exon.
PRDM7 is situated at the extreme tip of chromosome 16q, perhaps predisposing it to chromosomal copy number variation (segmental duplications to other other chromosomes), ironically mediated by meiotic recombination. The syntenic context TUBB3+ DEFB+ AFG3L1+ DBNDD1- GAS8+ PRDM7- qTel means PRDM7 is transcribed convergently with GAS8, a non-homologous conserved single copy gene whose distal exons are often detectable even in low coverage genomes in the contig containing PRDM7. This association has been extremely stable over placental mammal evolutionary time and so serves to reliably distinguish PRDM7 orthologs from its spin-offs.
The elephant genome has an old PRDM7 pseudogene adjacent to GAS8 in the expected opposite orientation. It has a second PRDM9-like copy in a novel syntenic location (ie unrelated to the CDH10-CDH12 location in primates) also seen in mammoth. Since afrotheres (plus xenarthrans) are the basal placental mammal, it follows that this locus too was spun off from PRDM7, establishing a 110 myr history of telomeric susceptibility of PRDM7 to repeated rearrangements.
Gene copy variation may be common in individual inheritance but a paralogous copy seldom becomes established across a species and even more rarely displaces the parental gene completely. Yet this scenario is repeatedly observed in evolutionary history of PRDM7. Thus the telomeric location of PRDM7 may predispose it to these events, but their persistance is not accidental: the erasure of meiotic recombination iniation sites by biased gene conversion drives evolution of PRDM7/9 at three different scales -- point mutation at key amino acid positions, expansion/contraction of zinc finger tandem repeat number, and whole gene copy number.
This history was not initially appreciated. Recall two genes in two species are orthologous only when they are vertically descended from the same gene in their last common ancestor which for human and elephant is post-marsupial mammal which had a single copy of PRDM7 adjacent to GAS8. The respective PRDM7 genes, still adjacent to GAS8 today are orthologous. The respective 'PRDM9' genes are however not descended from a common ancestral PRDM9 gene but from independent gene duplications of PRDM7 at different times during the course of afrothere and primate speciations.
Human PRDM9 lies in a retroposon-rich gene desert, flanked by two pairs of cadherin genes at a larger scale of 7 mbp. In rhesus, these same genes are seen (with some minor rearrangements), by parsimony establishing this PRDM9 segmental duplication preceded the divergence of old world monkeys.
Marmoset has a seemingly functional PRDM7 in the usual position facing GAS8, still at the extreme end of Callithrix chromosome 20. The cadherin cluster is intact on chr2:178,954,165-180,696,523. However Blastx of the intervening dna -- which is similar in size to rhesus and human so not suggestive of large deletions -- shows no suggestion of PRDM9. The assembly is gapless here. Blastx is sensitive enough to detect pseudogenes of this age provided they decayed only by small indels and nucleotide substitutions.
Thus PRDM7 had not yet duplicated in the primate stem placing that event just prior to old world monkeys/great apes divergence. Note that the marmoset PRDM7 has a respectable terminal zinc finger array of twelve units, enough to specify an adequately specific dna recognition sequence. Tarsier assembly has poor coverage and only a fragmentary PRDM7 gene presumed adjacent to GAS8.
Gene Strand Protein Start Species CDH18 - cadherin 18 19981287 homSap ponAbe macMul CDH12 - cadherin 12 22853731 homSap ponAbe macMul calJac PRDM9 + human PRDM9 23528704 homSap ponAbe macMul CDH10 - cadherin 10 24644911 homSap ponAbe macMul calJac CDH9 - cadherin 9 27038689 homSap ponAbe macMul
Lemurs present a new complication. The Otolemur assembly has two distinct and possibly functional PRDM7 copies with 8-11 zinc fingers, according to how distal stop codons and frameshifts are interpreted in low coverage assemblies. One of these lies in a contig AAQR03144890 also containing GAS8 end-sequence in expected opposite orientation but this copy of GAS8 is a segmentally duplicated pseudogene, representing a new type of lineage-specific larger segmental duplication. Authentic GAS8 lies in a different Otolemur contig AAQR03166494 lacking any sign of a zinc finger protein. The second PRDM7 gene lies in a contig AAQR03189271 with novel synteny to the gene ARFGEF1. There is no sign of primate PRDM9 (a homolog intercalated between cadherins CDH10 and CDH12).
The other lemur with an assembly, Microcebus murinus, has but a single presumptive PRDM7 with seven zinc fingers. The only relevant contigs (ABDC01433247 and ABDC01371462) contain no informative syntenic information so this gene cannot be associated with GAS8 with any confidence. However the contigs cannot be tiled and possibly belong to distinct genes.
The basal euarchtonal species, tree shrew Tupaia belangeri, has an unsatisfactory assembly. A putative PRDM7/9 gene can be put together utilizing raw traces reads located with lemur blastn queries. These cannot be convincingly tiled and thus could originate from multiple genes including related chimeric domain proteins, even though best reciprocal blast of each exon calls up established PRDM7/9 matches.
Moving on to laurasiatheres, Bos taurus presents a much more complicated situation. First, the GAS8 locus on chr18 contains the first two exons of a PRDM7 pseudogene in expected orientation but distal regions of the gene are completely deleted. The cadherin locus on chr20 is also intact but the 2.6 mbp region between CDH12 and CDH10 contains no indication of PRDM9, consistent with that segmental duplication being primate-specific and PRDM7 being the older parental location. This holds in the Baylor 4.0 assembly carried at UCSC, the Baylor 4.2 assembly, and the alternative assembly of the same data, UMD3.1. The latter two can be queried by the genomic blast server at NCBI.
A third locus on chr 1 hosts an unreviewed GenBank pipeline entry called PRDM9, derived as NW_003053109 from the alternative bovine assembly UMD3.1. NCBI staff corrected an unspecified frameshift to fix the reading frame -- a dangerous practice in a gene family prone to pseudogenization. The gene, called PRDM9a here, resides on the extreme end of chromosome 1 and differs from the Baylor 4.0 assembly at two amino acids outside the zinc finger region. The syntenic context here is novel: EFHB- RAB5A+ PCAF+ ZNF596- PRDM9a- which corresponds overall to human chr 3. The juxtapositioning of two zinc finger proteins on the same strand causes PRDM9 alignments to extend spuriously into the 12 zinc fingers of ZNF596, jumping over its 5 earlier coding exons.
ZNF596 contains a KRAB domain but no PR(SET) methylase. Humans encode a best-blast protein with the same assigned name on chr 8 (77% identity). Note the early exons of ZNF596 can be added to end of PRDM9a to form an artificial probe for this association in other species, though the two genes have a 43,400 bp spacer in cow, which is large relative to contig size in low coverage assemblies. The sole fragmentary transcript from yak testis (EF432551) is nearly identical to this PRDM9a, suggesting that the gene -- and perhaps its syntenic location -- became established prior to yak-cow divergence at 2.5 myr and is still functional. Its array of seven zinc fingers could recognize at most a region of 21 bp.
ZNF596 did not arise from a PRDM9-like gene through loss of the SET domain, though it is one of the better matches within the large zinc finger family. Excluding the zinc finger domain, ZNF343, ZNF133 and ZNF169 provide much higher blastp scores, as they also do just comparing the zinc finger arrays. The juxtaposition of ZNF596 and PRDM9a is likely coincidental rather than a consequence of inhomogeneous recombination between zinc fingers bringing PRDM9 to this site.
The fourth PRDM9 locus of interest, called here PRDM9b, is still not mapped to any bovine chromosome. It resides in contig DAAA02065087 in the UMD3.1 assembly and is temporarily assigned to chr Un.004.649 at Baylor assembly. Here the reading frame in exon two can be restored if a run of 5 A's is corrected to 6 A's, as homopolymer run length error is common in assemblies. The protein has a full set of domains KRAB SSXRD SET C2H2 with a moderate zinc finger array of five. Synteny cannot be determined in chr Un features which simple abuts unplaceable contigs into a manageable unit. Flanking dna in DAAA02065087 maps to several places in the cow genome, suggesting this feature has copy number attributes, perhaps of telomeric repeat type. PRDM9b is not a recent feature (or assembly stutte artifact) because it differs at toom any amino acids from other PRDM9 features in the cow genome. These substitutions avoid highly conserved residues, not consistent with cryptic pseudogenization. PRDM9b is capable of histone markup but it is not clear whether it does so.
Yet another locus in the Baylor 4.0 assembly, called PRDM9c here, could not initially be placed on a cow chromosome. While such features are often assembly artifacts, this one is supported by a transcript from 4-cell embryos (GO353654) consistent with a role in or after meiosis. In UMD3.1, this gene has been placed on chr X. Despite a very large contig, no zinc fingers occur in any reading frame, suggesting that the gene was transferred here without the last exon or it subsequently got deleted. In any event, the penultimate exon does not have a phase 1 splice donor in expected position and so would terminate at the next stop codon downstream. The protein retains the KRAB, SSXRD and SET domains but does not possess the ability to scan or bind dna. It has accrued various amino acid substitutions relative to other bovine PRDM9 that rule out recent establishment.
Finally, two additional genes, denoted PRDM9d and PRDM9e, are located as a parallel tandem pair in a higher quality region of bovine chr X. These are 96% identical as proteins, consistent with one being derived fairly recently from the other. Synteny here will not be informative until other ruminant genomes become available.
Overall the situation in cow is very different from primates and rodents. Results there about the function of single-copy autosomal PRDM9 genes in meiosis markup can scarcely be carried over to a species with five seemingly intact genes, three of which are on chr X. Ruminants have a well-characterized small pseudoautosomal region on which crossovers with chrY can occur.
The cow situation cannot be limited to the Hereford breed used for the genome project because the PRDM9 are too diverged from one another outside the zinc finger region. Indeed there is some suggestion from non-NCBI sheep genome that it too has many of these copies. Muntjak too seems similar to cow. However other cetartiodactyl genomes (dolphin, pig and alpaca) and other laurasiatheres (panda, dog, cat, shrew, bats) are not so expanded, suggesting that this unprecedented complexity could be limited to pecoran ruminants. The PRDM7 pseudogene is presumably parental to all these ruminant genes based on other laurasiatheres and placentals overall.
All-vs-all blastp percent identities are consistent with this, though rates of evolution in this gene family hardly fit standard paradigms for quantification. Results for bovine are summarized in the table below:
Gene #ZNF Status Chr Synteny cDNA Accession 9a_bosTau 9b_bosTau 9e_bosTau 9a_oviAri 9a_turTru 7_ailMel PRDM7 - pseudo 18 GAS8 no none -- -- -- -- -- -- PRDM9a 7 ok 1 ZNF596 yes NW_003053109 100% 85% 81% 82% 76% 72% PRDM9b 5 ok ? not det no DAAA02065087 81% 100% 78% 79% 72% 68% PRDM9c 0 ok X not det yes XM_002699750 80% 80% 82% 83% 74% 73% PRDM9d 9 ok X --- no none 80% 78% 96% 93% 73% 67% PRDM9e 9 ok X --- no none 81% 78% 100% 93% 73% 68%
Human PRDM9 variation
A great deal of attention -- and rightly so -- has been expended on cataloging variation in the zinc finger array at the level of both individuals and populations. While not the whole story of PRDM9 functionality by any means, this region is the primary determinant of recombination hotspot locations in meiotic dna. These sites greatly influence observed haplotypes and so the zinc finger array and its changing specificity over time must be understood to make reliable inferences about recent human evolutionary history and indeed speciation.
The zinc finger array is roughly analogous to tRNA. Both bind trinucleotides, the former in double-stranded dna and the latter in single-stranded messenger rna. Both are somewhat fuzzy in binding specificity, the zinc fingers only partly specifying a sequence (eg CCNCCNTNNCCNC) and tRNA accepting wobble codons. Both require an array, these are covalently joined and consecutive in the zinc finger array but are discrete and sequentially acting in tRNAs.
However this analogy only goes so far: the anticodons of tRNA have been fixed for billions of years whereas the four amino acid 'anticodons' of PRDM9 zinc fingers must undergo very rapid but highly restrictive mutation to keep up with an ever-changing recognition site (which obliterates itself with gene conversion, often the outcome of double-stranded break repair instead of recombination). Further, while all tRNAs recognize at least one codon, only a fraction of the zinc fingers in the human PRDM9 array can be utilized -- 13 fingers specify 39 nucleotides whereas observed sites are far shorter, some 13-17 base pairs. What selective pressure then maintains the unused fingers?
That is but one of many remaining questions about PRDM9. Expression in some mammals is not restricted to germ line cells, suggesting other functionalities in the regulation of gene expression. The PRDM9 locus on chr5 itself does not contain a notable recombination hotspot (relative to its own zinc finger array) so gene conversion here cannot explain its mutational frequency, focus on the four determinative residues, and restricted compositional outcome (to nine of twenty amino acids).
Selectional pressure on this gene is highly unusual in that an amino acid substitution in a germline cell yielding a zinc finger that cannot recognize a meiotic target is eliminated right away because recombination is essential to the meiotic process, meaning that no correctly divided haploid cell is available for fertilization. Other regions of the same protein evolve much more conventionally, with human PRDM9 diverging overall from other primates at unremarkable rates.
The zinc finger array varies not only pointwise but also in number of repeats, from 13 or fewer to 20 or more, in contrast to many other stable 'polydactylic' zinc finger proteins. The mutational mechanism by which repeat numbers contract and expand has not been established but is presumably replication slippage, as in other unrelated proteins (such as the octapeptide repeat region in human PRNP). It is unclear what happens to individual zinc finger utilization after an expansion or contraction.
Note in males, recombination must occur in the two short pseudoautosomal regions of homology between chrX and chrY where few base pairs are available (relative to much longer autosomal chromosomes) for the recognition sequence to occur randomly with reasonable probability. Thus in humans PAR1 on the short-arm ends of chrX and chrY is 2.6 mbp whereas as PAR2 on the long arms ends only comprises 320 kbp. By comparison, the shortest human chromosome, chr22, has 50 million bases to host recombination recognition sites (16x as much). Thus the PARs may provide the do-or-die selectional bottleneck driving zinc finger array evolution.
Given that small surveys in moderately inbred populations (such as Iceland) already find considerable variation in both number and sequence particulars of PRDM9 zinc finger arrays, it seems inevitable that many individuals must be heterozygous, sometimes radically so. However these would not necessarily be reported from sequencing projects where commonly only one allele is determined. It is not known whether both alleles in a heterozygous individual would be expressed and participate on an equal footing in meiosis in the same dividing cell. If so, the repertoire of recognizable sites would be expanded, with complications for understanding haplotype evolution if common.
One last immense complication is that human and mouse do not speak for the rest of mammals. There, multiple copies are present in some major lineages, in some cases with zinc finger arrays too short to determine an adequately restrictive suite of recombination sites. Here the possibility must be considered that paralogous copies can act in tandem with short arrays acting in concert to define adequate length sites. The pseudoautosomal regions are by no means strictly conserved phylogenetically. Here adequate data may well be available from horse and cattle breeders but it has not surfaced to date.
The role of CpG mutations
Human PRDM9 has 39 CpG sites in its coding exons, potentially methylated on the C, subject to spontaneous deamination to uracil and mis-repair, and so mutational hotspots. After attempted dna repair, the resulting change can be either CpA or TpG. These changes alter the encoded amino acid at non-synonymous sites. Some 28 of the CpG sites of PRDM9 are at arginine CGn codons (of which the protein has 90 overall).
These always result in a substitution: G -> A mis-repair yields histidine for CGT and CGC and glutamine for CGG and CGA; C -> T mis-repair leads to cysteine for CGT and CGC and tryptophan and stop codon for CGG and CGA. These changes indeed occur in reported human and mammal sequences where they are perhaps best viewed as cSNPs in an individual rather than representing the species as a whole. The display below shows wildtype human PRDM9 in the top lines and the effects of G -> A and C -> T in the next.
In terms of upstream CpG islands that would protect against methylation of CpG in coding regions, PRDM9 has none. While three occur somewhat near the start of PRDM7, these do not extend into coding exons and may not even be associated with this gene. The composite snapshot below from chr5 and chr16 of the UCSC human genome browser displays these CpG islands relative to the two genes. Thus CpG cytidines would be methylated in coding regions of both PRDM7 and PRDM9, rendering them susceptible to hotspot mutations.
In the terminal zinc finger array of the human PRDM9 reference sequence, position -1 is sensitive to the CpG hotspot effect. However rapid rapid evolution in the zinc finger array, which is overwhelmingly concentrated in the four dna-recognizing residues, cannot be explained by the CpG effect. On the other hand, the common alteration of the terminal partial finger YVCREDE* to Y*CREDE* in some species likely is a CpG effect but one that is insufficient for loss of function.
PRDM9_homSapWT MSPEKSQEESPEEDTERTERKPMVKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITIGLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQVKPPWMALRVEQRKHQKGMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKLELRKKETERKM
PRDM9_homSapCA ...................Q.............................H...................Q......Q...................................H...................................................................
PRDM9_homSapTG ...................W.............................C...................*......*...................................C........V..........................................................
PRDM9_homSapWT YSLRERKGHAYKEVSEPQDDDYLYCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWLITKGRNCYEYVDGKDKSWANWMRYVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDE
PRDM9_homSapCA ...Q...........K........................................H.........................................Q....K......................................Q.....................Q...............
PRDM9_homSapTG ...*............L.......................................C..............................L..........*...........................................W.....................*...............
PRDM9_homSapWT YGQELGIKWGSKWKKELMAGREPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK
PRDM9_homSapCA .S..............................................H.....................................H............................................................................
PRDM9_homSapTG ................................................C.....................................C............................................................................
........-1..23..6.......... ........-1..23..6.......... ........-1..23..6.......... ........-1..23..6..........
VKYGECGQGSVKSDVITHQRTHTGEKL YVCRECGRGSRQSVLLTHQRRHTGEKP YVCRECGRGRDKSHLLRHQRTHTGEKP
........................... .......Q..Q................ .......Q.HN................
........................... .......W..W................ .......W.C.................
YVCRECGRGSWKSHLLIHQRIHTGEKP YVCRECGRGSWQSVLLTHQRTHTGEKP YVCRECGRGRDKSNLLSHQRTHTGEKP
.I.....Q................... .......Q................... .......Q...................
.......W................... .......W................... .......W...................
YVCRECGRGSWQSVLLTHQRTHTGEKP YVCRECGRGSWQSVLLTHQRTHTGEKP YVCRECGRGSNKSHLLRHQRTHTGEKP
.......Q................... .......Q................... .......Q...................
.......W................... .......W................... .......W...................
YVCRECGRGSRQSVLLTHQRRHTGEKP YVCRECGRGSNKSHLLRHQRTHTGEKP YVCRECGRGRNKSHLLRHQRTHTGEKP YVCRECGRGSDRSSLCYHQRTHTGEKP YVCREDE
.......Q..Q................ .......Q................... .......Q.H................. .I.....Q..N................ .I.....
.......W..W................ .......W................... .......W.C................. .......W................... .......
A weblogo based on alignment of placental mammal PRDM7 and PRDM9 genes (with pseudogenes excluded) illustrates the location of expected CpG mutations relative to conserved residues. These will be relatively high frequency loss-of-function alleles (not affecting health per se if only reproductive meiosis is affected).
In the initial KRAB domain, the potentially affected arginines are not especially well-conserved. However, at the first site, neither histidine nor cysteine is part of the reduced alphabet so these changes are unlikely to be tolerated in meiotic functioning. At the second and third sites, glutamine does occur secondarily in some species (cow, sheep and muntjac) and murid rodents, respectively. These changes are thus borderline for adverse effects on functionality.
Sequence analysis of human variation
The PRDM9 terminal zinc finger array varies extensively in human, with significant consequences for hotspot recognition motif, distribution of recombination location options along the chromosomes, population history (linkage disequilibrium), and chromosomal rearrangement diseases. No other species -- notably other great apes -- has been surveyed to any extent for individual variation (with the exception of mouse PRDM7 where hybrid sterility was first mapped).
For these species, we have only the sequence of the animal selected for genome sequencing and so have no idea whether human variation is unique or typical. With high priority chimp, Genbank contains only an uncurated erroneous gene prediction XM_517829 and an array fragment GU166820 with a disturbing number of differences to chimp reference genome. Gorilla is worse. Mouse has considerable variation in its zinc finger array but the strains involved are highly inbred and not necessarily representative of wild mouse diversity.
Cheap short reads mapped to human reference as SNPs prove highly unsatisfactory for genes like PRDM9 where individuals differ not only at pointwise sites but also in wholesale repeat number. Several labs have reported novel repeat multiples but found an hour of re-sequencing too tedious; others assumed all possible arrays had already been reported and forced reads into one of these pre-existing classes; others left their discoveries as article graphics, behind firewall or in supplemental, not troubling themselves with GenBank entries, with laudable exceptions. Even if certain arrays are rare, they provide invaluable information on the genetic mechanisms by which repeat number variation arises.
It appears that few individual human genome or exome projects really gathered enough data to allow ab initio assembly of the zinc finger repeat array, or even when they did, walked away from that exercise, deposited a mess of indels and base miscalls at the Short Read Archive and then claimed SNPs relative to human reference, contaminating that resource with error.
This is very unfortunate in the case of both basal and ancient human dna, which might record intermediate or population-specific stages in the evolution of human PRDM9. Extracting accurate bushman, paleo-eskimo, neanderthal, and denisova PRDM9 zinc finger arrays requires starting from scratch from raw read data. This may however be impossible due to inadequate coverage and confusion of short reads with PRDM7 and even within PRDM9, not to mention other closely related zinc finger proteins.
Here PSU provides an excellent display of reads (along with quality scores) reported by the various projects. The final exon of PRDM9 can be viewed (noting PSU uses hg18 coordinates) at chr5:23562098-23562523 for the early region and chr5:23562524-23563636 for the terminal zinc finger array. Viewing the display to dense mode shows the extent of tiling: it does not appear that adequate coverage was obtained in the critical cases. Here it cannot be assumed that the zinc finger array of bushman (who represent the earliest diverging living relative of Europeans) will closely resemble extensively sequenced West African variants (Yoriba). The best that can currently be done with bushman genome is VKYGECGQGFSVKSDVITHQRTHTGEKL YVCRECGRGFSWKSHLLIHQRIH ... YHQRTHTGEKP YVCREDE* which matches hg19 human reference sequence without shedding any light on internal repeat length or sequence variation.
Although the zinc finger array conveniently resides in a single exon, that exon is almost never sequenced in its entirety. It has never been sequenced as a byproduct of an expression project. Consequently we have no idea its early zinc finger covaries with the terminal array nor any understanding of the constraints acting on the long bridging domain.
10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170
| | | | | | | | | | | | | | | | |
PRDM9_homSap EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAKVKYGECGQGFSVKSDVITHQRTHTGEKL
PRDM9_panTro ...............................................................R................................................................A........................D.............G.P
PRDM9_gorGor .........................................T.....................R.........................................................................................................P
PRDM9_ponAbe .......................................................H....S..R.C.......................................................................................D.............GRS
PRDM9_nomLeu ...A......................A.H.............F.................S..R.C...............................S..V...........I..-.............Q...........E............................
PRDM9_macMul ...............................T.........R.F....L.S.........S..R.C.....................PK................S...E.M....Y........E...........................D.....I.........P
PRDM9_papHam ...............................T...R.....R......L.S.........S..R.C....................R.K................S...E.M...........S.E.I.........................D....VI.........P
PRDM7_calJac .S.....................H.............T.T............K.KE....F..CNS........T........I......MA....N........S..EE.M.............VD...............A..........DM...TG.........P
PRDM7_micMur ............S.............KHT....IS.RT.G..H................HS....C...A.D..V...P.PFH.K.Q..G...........K.....E........P......G..D.D..CAAG.....SR....DS..S..D..N..I.........P
PRDM7_otoGar ............S....T..........T.P..ISQ.T.G..N.R.QT...R.E.....HS..N..........V..M..TSH.K.Q.SR...I..C........S.E.E.MI...P.PD...G..D.E.FC.AI...G.V...NR..V.S..N..N-LR.........P
A terrible error was made early on in human PRDM9 variant nomenclature. It is completely unacceptable for PRDM9 to have a private system for naming zinc fingers that stands in conflict with dozens of previously established crystallographic structures, SwissProt, SCOP and PFAM practice, and nomenclature for the other 842 zinc fingers encoded in the human genome.
Wrong as it is, the current nomenclature will not be easy to displace. However this site uses a nomenclature consistent with physical structure, comparative genomics and historical precedent throughout and only provides a partial translation table to the numerous misguided motif naming systems.
The mistake arose because the first and last zinc fingers in primate PRDM9 are mildly anomalous. However it is exceedingly common even for internal zinc fingers to depart from canonical form, even to admit different spacings and substitutions in the C2H2 ligands, as well depart in length and cap domain. Zinc finger arrays commonly terminate in fragmentary motifs that often continue for a while in another reading frame (ie, represent non-3n indels with run-on to the next encountered stop codon).
Even when each zinc finger is letter-perfect, only a small subset seem to function in dna recognition -- thus 15 zinc fingers in a PRDM9 variant could theoretically recognize a 45 bp dna sequence but a look at meiotic events show that 17 bp seems the upper limit for specificity, meaning no more than 6-7 motifs are utilized in vivo. Nomenclature must acknowledge all zinc fingers whether they are anomalous or not (or functional or not).
Some 7,000 of the overall ten thousand zinc fingers end in a structurally distinct cap unit, typically TGEKP. This was shown long ago to lock the zinc finger down after scanning and has found the recognition sequence. Proteins with a single zinc finger still have this motif. It occurs at the end -- not the beginning -- of the main zinc binding region. Proline is no accident here: as a cyclic imino acid, it is structurally terminating for helix and sheet.
In summary, zinc fingers begin 5 residues before the second zinc cysteine, not at the second cysteine as in the 'ABCD...' nomenclature. Human PRDM9 begins with a full length zinc finger, but with a lysine at position 2 replacing the usual branched aliphatic, a tyrosine at position 3 replacing the first cysteine and a leucine replacing the terminal proline: VKYGECGQGFSVKSDVITHQRTHTGEKL. These oddities became stably established in the theran ancestor 135 myr ago (though departures -- and even the expected residues -- are seen in some species). Otherwise, the first zinc finger is quite conventional. The terminal leucine surprisingly is seen in all reported human variants. While the first zinc finger assuredly has the zinc finger fold and likely binds zinc to some extent, it likely does not function directly in specific dna motif recognition. Its role may more that of a macro cap, facilitating the lineup of downstream zinc fingers.
Similarly, the terminal YVCREDE* fragment, with its anomalous charged aspartate and glutamate in place of cysteine and glycine, is not zinc binding or part of a dna recognizing motif but simply a partial end cap. It has persisted (imperfectly) since the boreoeutheran ancestor so evidently provides significant value. In many laurasiatheres it is YCRECE or even YRCREG. Even a canonical hexapeptide cannot reach across the preceding TGEKP cap to displace zinc binding residues of the last full repeat, nor can the six residues circle around to displace the first five residues of first repeat. Instead, these residues form the start of an additional fold, enough to that keep the repeat array from unraveling.
Below the available diversity at GenBank is shown, with the 0th GEKL repeat removed because it has no variation. Also the longest allele has its last two repeats removed to shorten the display width. The terminal fragment is also not shown. Redundant sequences have been largely removed from the set of 42. The alignment is shown at the protein level because synonymous dna variation is largely irrelevant to function.
If all 42 human variant zinc finger array alleles at GenBank are collected, parsed into their zinc fingers and aligned for their differences relative to the genomic reference sequence, 25 variant fingers emerge at varying frequencies of occurrences. These are provided below, ordered by subgroup. The full sequence, allele name of the original investigators and representative accession are also given.
As previously observed, variation at the amino acid level is overwhelmingly concentrated at that handful of internal positions recognizing dna bases in the major groove. Furthermore, the amino acid substitutions are strongly concentrated within only 9 of the 20 available amino acids. These observations raise the question of how ordinary random mutational processes could possibly have produced these results. Perhaps variation elsewhere results in failed meiosis, causing these to disappear immediately, leaving only the observed variation.
Note in the table below (whose underlying data is here) that threonine appears as a very common alternative to isoleucine outside the critical region (between the two zinc-binding histidines): YVCRECGRGFSWKSHLLIHQRIHTGEKP. This is actually an oddity of the first GEKP repeat: T is found here in all other primates (ie ancestrally), not I. As bushmen also have I here, this allele was fixed prior to their divergence at 70,000 years.
Human Variation in 507 Zinc Fingers in 42 PRDM9 Variants
Difference to NM refSeq Freq Full length zinc finger Accession Allele Great Ape Zinc Finger Variation
YVCRECGRGFSWKSHLLIHQRIHTGEKP 39 YVCRECGRGFSWKSHLLIHQRIHTGEKP NM_020227 ref YVCRECGRGFSWKSHLLIHQRIHTGEKP homSap
......................R..... 1 YVCRECGRGFSWKSHLLIHQRIRTGEKP FJ899869 7 .................S...T...... 1 panTro
............Q.V..T...T...... 100 YVCRECGRGFSWQSVLLTHQRTHTGEKP NM_020227 ref ...........V..S..S...T...... 6 panTro
............Q.V..S...T...... 22 YVCRECGRGFSWQSVLLSHQRTHTGEKP GU216222 A ...........V..S..S.RTT...... 1 panTro
............Q.V..R...T...... 13 YVCRECGRGFSWQSVLLRHQRTHTGEKP GU183919 CH3 ...........VQ.N..S...T.....L 1 panTro
......R.....Q.V..T...T...... 1 YVCRECRRGFSWQSVLLTHQRTHTGEKP HM211000 L18 ...........QQ.N..S...T...... 1 panTro
............Q.VP.T...T...... 1 YVCRECGRGFSWQSVPLTHQRTHTGEKP FJ899895 18a ...........RQ.A......T...... 1 panTro
...........N.....R...T...... 65 YVCRECGRGFSNKSHLLRHQRTHTGEKP NM_020227 ref ......E....QQ....R...T...... 1 panTro
..........RN.....R...T...... 39 YVCRECGRGFRNKSHLLRHQRTHTGEKP NM_020227 ref ...........QQ....R...T...... 2 panTro
..........RK.....R...T...... 1 YVCRECGRGFRKKSHLLRHQRTHTGEKP GU183915 AA2 ...........QQ....S...T...... 2 panTro
..........RD.....S...T...... 14 YVCRECGRGFRDKSHLLSHQRTHTGEKP GU216229 I ...........KQ....S...T...... 2 panTro
..........RD.....R...T...... 27 YVCRECGRGFRDKSHLLRHQRTHTGEKP NM_020227 ref ...........RQ.V......T...... 1 ponAbe
..........RD..N..S...T...... 48 YVCRECGRGFRDKSNLLSHQRTHTGEKP NM_020227 ref ...........RR.V......T...... 1 ponAbe
..........RD..P..S...T...... 1 YVCRECGRGFRDKSPLLSHQRTHTGEKP GU183915 AA2 ...........QQ.V......T...... 1 ponAbe
..........RD..N..S...T...D.. 4 YVCRECGRGFRDKSNLLSHQRTHTGDKP GU183915 AA2 ...........RR.V......T...... 1 ponAbe
..........RDE.N..S...T...... 2 YVCRECGRGFRDESNLLSHQRTHTGEKP HM211006 24L ..............V..R...T...... 1 ponAbe
..........RDQ....S...T...... 1 YVCRECGRGFRDQSHLLSHQRTHTGEKP GU183919 CH3 ...........QQ.VVF....T...... 1 ponAbe
...........RQ.V..T...T...... 2 YVCRECGRGFSRQSVLLTHQRTHTGEKP FJ899905 10b ...........G..V.FR...T...... 1 ponAbe
...........RQ.V..T...R...... 79 YVCRECGRGFSRQSVLLTHQRRHTGEKP NM_020227 ref ...........D..GVCY...T...... 1 ponAbe
...........RQ.V..T...G...... 2 YVCRECGRGFSRQSVLLTHQRGHTGEKP FJ899872 10 ...........V..N..S...T..E..L 1 ponAbe
...........RQ.V..S...T...... 1 YVCRECGRGFSRQSVLLSHQRTHTGEKP GU216228 H ...........D..S..R...T...... 3 nomLeu
...........NQ.V..T...T...... 1 YVCRECGRGFSNQSVLLTHQRTHTGEKP GU183916 AA11 ...........K..N..S...T...... 1 nomLeu
...........DQ.V..T...T...... 1 YVCRECGRGFSDQSVLLTHQRTHTGEKP GU183916 AA11 ...........V..N..S...T...... 1 nomLeu
...........DR.S.CY...T...... 37 YVCRECGRGFSDRSSLCYHQRTHTGEKP HM210983 L1 ...........Q..S..S...T...... 3 nomLeu
...........DR.S.CY...T..MSKS 5 YVCRECGRGFSDRSSLCYHQRTHTMSKS GU183916 AA11 .L.........V..S..S...T...... 1 nomLeu
507
When the 42 variants are aligned at the dna level, synonymous variation might be anticipated more or less evenly across the repeat array under the assumption that natural selection acts here only at amino acid level. However this is not the case as shown in the graphic below. For example, the GEKL repeat has no variation whatsoever despite numerous 4N codons. Elsewhere, synonymous variation is again highly concentrated at residues important to meiotic repeat recognition. This suggests a novel mutational mechanism exists that focuses change at the key regions, at a rate far above the genomic average. Conceivably the dna itself might have additional hairpin structure that exposes the critical regions to enhanced mutation. Such a speculative structure would fit with replication slippage varying the number of array repeats. This mechanism can also sweep out variation. Alternatively, the observed distribution of synonymous variation could arise via hypothetical mRNA editing in conjunction with a retroposon-like or copy-editing mechanism. A third option envisions another protein recognizing the dna encoding the repeats and acting upon them to provide variation.
The terminal zinc finger array in the human reference sequence (but not chimp or orangutan) has a two-block structure. That is, the repeats 3-7.5 have a high degree of internal self-similarity as do repeats 7.6-13.4. However these two blocks are markedly dissimilar to each other, primarily due to transversions (rather than transitions C<->T or A<->G). The genetic code is such that transversions give rise to markedly less conservative amino acid substitutions in both physical and dna binding properties, as can be readily seen at the protein level for human PRDM9.
The origin of the two-block feature cannot be dated accurately because of limited sampling of individuals in great apes but is likely specific to human (or even to some human populations). It is closely correlated with the history of repeat contraction and expansion in this rapidly changing region of the gene. Note the zinc finger array is neither a microsatellite being 84 bp long vs the requirement of 1-6 bp nor a minisatellite.
Dotplots can create visual artifacts, depending on scanning window and mismatch settings. Here the two-block structure is highly robust to exploration of parameter space and its basis is readily apparent at both the dna and protein sequence levels.

A very recent PNAS article looks at many human meioses and assigns the recognition sequence to the distal region of the zinc finger array (in Fig 1D and supplemental S2), which corresponds to the second block identified above. That raises the question whether the two blocks evolve by different non-mixing mutational mechanisms and leaves unexplained the functional tasks implied by observed conservation of the first block. The lack of block structure in chimpanzee PRDM9 illustrates once again that meiotic initiation is evolving in many different directions.
Rate of proximal PRDM9 evolution in primates
The rate of evolution of human PRDM9 at the protein level -- excluding special evolution in the terminal zinc array -- is rapid but perhaps not unusually so. A rate anomaly can only be defined relative to the rather skewed rate distribution of the human proteome (20,000 loci). However a better comparison might be just to rates of the many other KRAB, SSXRD and PR(SET) domains in the proteome and to linker regions which are often under little selection. These regions have mediocre conservation in general and so a rapid rate there for PRDM9 has no immediate implications for its association with meiosis or protein binding partners.
The fact that PRDM9 is a recent gene duplicate of PRDM7 adds another rate complication, as gene duplicates often exhibit rapid initial evolution as the copies subfunctionalize, a problem exacerbated here by functional persistence of a variously truncated PRDM7 in some primate lineages. Independent duplications of PRDM7 (eg afrotheres and pecorans) further complicate rate considerations outside of primates.
Together, these considerations make it difficult to define a meaningful 'peer group' in any major clade by which to benchmark the rate of PRDM7/9 evolution. However by any measure this gene family is not evolving slowly in placental mammals, perhaps surprising because excluding the zinc finger array leaves domains with seemingly fixed and demanding functionality. That is, the KRAB domain is co-evolving with its protein binding partners which are under many other constraints. The histone substrate for methylation is exceedingly conserved but the PR(SET) catalytic domain of PRDM9 is not, despite its narrow specificity.
The difference alignment below shows change localization relative to human functional domains within other primate PRDM9. The comparison includes catarrhine PRDM7 corrected where needed for frameshifts and stop codons as well as PRDM7 from euarchontal species that diverged before the gene duplication. The 532 residues are relatively free of deletions or insertions.
None of the four sites where human diverges from long-established consensus (R5K, P155S, G178R, R445H) are CpG hotspots. With the exception of G178R, these are conservative substitutions in linker regions and likely near-neutral. G178R represents a radical change in amino acid properties within the SSXRD domain. However this is not a conserved residue in the parental SSX1 domain.
Sequencing accuracy is an issue here because some genes are missing exons altogether and other exons have only single trace coverage. Outside of human and mouse, only a single individual has been sequenced. However humans are not especially variable in the proximal region of the gene, only a single coding SNP is known to date (R113C), in marked contrast to the zinc finger array. Much more intensive sequencing of primates is essential to quantitative understanding recent evolution or PRMD9 -- the 16 species sampled so far represent only 5% of living primate diversity. For example, the flying lemur divergence node is not represented at all.
<------------------------ KRAB domain ------------------------> cSNP:R113C <-------- SSXRD domain ---------><-------- zinc knuckle ----> <---------------
PRDM9_homSap MSPEKSQEESPEEDTERTERKPMVKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITIGLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQVKPPWMALRVEQRKHQKGMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKLELRKKETERKMYSLRERKGHAYKEVSEPQDDDYLYCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWN
PRDM9_panTro ....R...................................................................................................L.................................................P................K.....G....................................................................................
PRDM9_gorGor ....R...................................................................................................C.................................................P......................G............................................I.......................................
PRDM9_ponAbe ....R.......D.........T.....................................................................................................N.........E...................P.................S....GNT.............I.........-....................................T.....................
PRDM9_nomLeu ....R..............Q..T......................................................................................M........................GA..................P.................R....G.....*............................T...........I...T.G...............................
PRDM9_macMul ....R.................T....................................................................................V....S...........N.......V.GM.....T............P...R.............R....G..............................................I.....E...............................
PRDM9_papHam ----------------------------------------------------------------------------------------------------.......F....S....E...T............G.P...ST.........A..P.................R..A.G..................L.................................N...............................
PRDM7_homSap ....R.......G..........................................M.......V...........................................F.G..S...........N.....R...G.P....T.D..........P.................R....G...............I....................................................................
PRDM7_panTro ....R...................................................................................................L.................................................P................K.....G....................................................................................
PRDM7_gorGor ....R.......G........................................................Q.V............................-----------------.......N.........G.P....T............P...........R.....R....G...............I.K....................................R.............................
PRDM7_ponAbe ....R......KG...........................T.................KT...............................................F.G..S...........N.........G.Q....T............P..........T..I...R....G.T..................................................................................
PRDM7_nomLeu ----------------------------------------------------------------------------------------------------I.S....V....S...........N...G.....GSQ....T..*...R.....P...........Q.....R....G.....*............................T.......................H.........................
PRDM7_macMul ----------------------------------------------------------------------------------------------------.......F....S....E...T..N.........G.P...ST.D..*....A..P.................R....G.........R.....A..L.H............................N..N...............................
PRDM7_papHam ....R..............W.......................................................................................V....S...........N.......V.GM.....T............P...R.............R....G..............................................I.....E...............................
PRDM7_calJac ....R.......G..G...Q............M..S.................M..................................................G..F..G.S...........G......K..G...V..T..P.........P.................R.D..E..........L.................I.............................HA........................
PRDM7_tarSyr ...DR.P.D...G..G...C.SA.........................I..........T..A.....P........KR...PL.......................F....N.......R.PL.IV.......EM....^T.D....W......^.....E....K.I.F....I.VN........DC.....N...........Q..........T..I...IN....................................
PRDM7_micMur ...N........V.AG..GW..TD...........S.....Q......I..........V........P.............H.................----------------------------------------------------------------------------------------------............K.......................................K.R.............
PRDM7_otoGar ----------------------------------------------------------------....P...........T.YK..................H....F.M..S.R..ILK.CML.FNMH.....GP.S.P.I.....H..HM.SPR.........GR.SD..I..I.VR...........................K.......N..V.........T..E.......V....S..G.RT.......F....
---------------------------- PR(SET) domain -------------------------------------------------------> <- early zinc finger -->
PRDM9_homSap EASDLPLGLHFGPYEGRITEDEEAANNGYSWLITKGRNCYEYVDGKDKSWANWMRYVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIkWGSKWKKELMAGREPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK
PRDM9_panTro ..............K.......................................................................................................................................................................R................................................................A.............
PRDM9_gorGor ..............K.................................................................................................................................................T.....................R..............................................................................
PRDM9_ponAbe ...................K..........................................................................................................................................................H....S..R.C............................................................................
PRDM9_nomLeu ..................................................................................Q.......P........................T.E....A......................A.H.............F.................S..R.C...............................S..V...........I................Q...........E
PRDM9_macMul ...................Q..................................................................................................................................T.........R.F....L.S.........S..R.C.....................PK................S...E.M....Y........E................
PRDM9_papHam .....................K.................................----------------------------------------------------------------...............................T.........R......L.S.........S..C.C......................K................S...E.M.............E.I.....E........
PRDM7_homSap ..........................S......................S............................................S.................................................................................R..S..RCC.......................................S...E.M..............................
PRDM7_panTro ..............K...................................................................................................................................................-.....*.......R..S..RCC........V.........W............L.......S...E.M..............................
PRDM7_gorGor .....................K....S...........................................L............................................T............................................................R..S..RCC..................^....^...............S...E.M..............................
PRDM7_ponAbe .....................K......................................W.......................................................P...........................................H...A..............S..DCC...............................SA......S...E.M.....G........................
PRDM7_nomLeu .................................................*....K.......H...................Q.......P........................T.E...............V.T..........C.................R..............S.SR.C...............-...I...K.......L.......S...E.M..............................
PRDM7_macMul .............C.......K....S........................K...----------------------------------------------------------------......Y................^....................S..............S..S.C.........................L...T.........S...E.M....F.....A............E......
PRDM7_papHam ...................Q.........................................................................................................Y......................................S..............S..S.C....................R..Q.L...T.........S...E.M.K..F.....A............E......
PRDM7_calJac .................V.......SS.............................................................................................S.....................H.............T.T............K.KE....F..CNS........T........I......MA....N........S..EE.M.............VD...............
PRDM7_tarSyr ...E............Q..D......S........................................................I....................................T..K.L..S..S..L.....F....KC..PP.I...T....YV.......E.L.....QS....W...S.C..A.....PMH...Q...S..SL.N..TE.TE.S.EKE.M.K..PS.S...HL.D..YE.HHI.A.AAR-
PRDM7_micMur ...E............QV........S....................D..............E...................Q.............................E..TIRQ............S.............KHT....IS.RT.G..H................HS....C...A.D..V...P.PFH.K.Q..G...........K.....E........P......G..D.D..CAAG.....SR
PRDM7_otoGar .....Q..........QV........S....................E.QG...........E....................................................T..Q............S....T..........T.P..ISQ.T.G..N.R.QT...R.E.....HS..N..........V..M..TSH.K.Q.SR...I..C........S.E.E.MI...P.PD...G..D.E.FC.AI...G.V-
As expected, percent identity declines monotonically with increasing time of divergence. The rate of amino acid substitution, subject to the caveats above, places PRDM9 in the lowest quartile of human protein conservation, but with thousands of proteins evolving still faster. Variability is noticeably but not exclusively concentrated in the linker between the KRAB and SSXRD domains and in the long terminal linker. Variation within the PR(SET) domain largely avoids deeply conserved residues defined from comparison of the 16 distinct PRDM genes and 35 additional SET domains in human.
The table below shows percent amino acid difference relative to human, first for the entire 523 residues preceding the array and then separately for the three domains. Note the KRAB and PR(SET) are evolving slower than the overall proximal region while the SSXRD domain is changing more rapidly (however it is short so subject to wide rate swings).
Missing exons were supplied here by merging incomplete sister taxa (lemurs) or taking them from the closest source (gibbon, gorilla, tree shrew PRDM7). Taken human/tree shrew divergence at 90 million years, roughly one substitution per million years has been occurring, much of it in the last exon between the early zinc finger and terminal array.
In summary, the proximal region of PRDM9 is evolving quite differently than the zinc finger array. It is difficult to distinguish here between adaptive change, neutral drift, and substitutions driven by the role of PRDM9 in meiosis and recombination.
KRAB SSXRD PR(SET) 100% 100% 100% 100% PRDM9_homSap Homo sapiens (human) 98% 100% 93% 99% PRDM9_panTro Pan troglodytes (chimp) 98% 100% 96% 99% PRDM9_gorGor Gorilla gorilla (gorilla) 96% 100% 84% 99% PRDM9_ponAbe Pongo abelii (orangutan) 94% 100% 90% 98% PRDM9_nomLeu Nomascus leucogenys (gibbon) 94% 100% 93% 99% PRDM9_macMul Macaca mulatta (rhesus) 94% 96% 90% 97% PRDM7_homSap Homo sapiens (human) 96% 100% 93% 99% PRDM7_panTro Pan troglodytes (chimp) 94% 96% 87% 97% PRDM7_gorGor Gorilla gorilla (gorilla fusion) 93% 95% 90% 98% PRDM7_ponAbe Pongo abelii (orangutan) 91% n/a 90% 95% PRDM7_nomLeu Nomascus leucogenys (gibbon fusion) 93% 100% 93% 99% PRDM7_papHam Papio hamadryas (baboon) 90% 95% 87% 97% PRDM7_calJac Callithrix jacchus (marmoset) 80% 87% 78% 95% PRDM7_tarSyr Tarsius syrichta (tarsier) 81% 90% 84% 92% PRDM7_micMur Microcebus murinus (lemur fusion) 73% 76% 75% 92% PRDM7_tupBel Tupaia belangeri (tree shrew)
Among the 843 human genetic loci encoding zinc fingers proteins, the arrays most closely resembling PRDM9 in length, structure and amino acid composition are ZNF133, HKR1, ZNF343, ZNF589, ZNF169, ZNF596. While the functions of these proteins are largely unknown, the first two have a KRAB domain, a spacer, early zinc finger in the terminal phase 2 exon, and a zinc finger array similar in size to human. The next two are similar but lack the spacer, with the KRAB domain encroaching into the final exon. The final two have only the KRAB domain and terminal array. Some 290 human gene products encode a KRAB domain.
Here ZNF133 and the misnamed HKR1 are the best candidates for donating (via inhomogeneous recombination) the zinc finger array to the nascent PRDM7 which was already a chimer of KRAB, SSXRD and PR(SET) domains. The relationships here might instead go the other way (domain loss in PRDM) but different intronation of the KRAB domain is incompatible with that scenario. While none of the six ZNF is capable of histone methylation, KRAB domains are capable of recruiting SETDB1, a H3K9 methylase, partnering with the TIF1ß co-repressor protein (encoded by TRIM28), which interacts with many KRAB domains).
Phylogenetic variation in the zinc finger arrays of these proteins is potentially quite informative, the question being whether their variation too is focused on the four amino acid positions providing dna binding specificity in PRDM7/9. This next sections examine each protein separately for mutational variation in the zinc fingers over placental mammal evolutionary time.
Here the 46-species genomic alignment at UCSC serves as initial source of zinc finger arrays, which are then tested by blat back into individual species and then parsed into separate fasta files for each protein finger (the formats needed by the Multalin2 variable width differential aligner, weblogo and DotPlot tool).
ZNF133 and HKR1
Human ZNF133 is a conventional KRAB-zinc finger array (that lacks however the PR(SET) domain). Although the KRAB domains are only 31% identical, the array provides a better model for PRDM9 than the other 14 PRDM* loci in terms of zinc repeat character and length. However rodents cannot be used here as a model system for ZNF133 as the mouse syntenic counterpart is a known pseudogene -- as is rat but not guinea pig or rabbit. ZNF133 is yet another protein in this class that does not readily track back into marsupials or earlier vertebrates.
As with PRDM7/9, the C-terminal run-off of ZNF133 is subject to frameshifts. However elephant and human are still 86% identical in their last exon, with zinc finger arrays even higher. Armadillo, another mammal diverging from human at 101 myr, is 91% identical in this region and has exactly the same number of zinc fingers (14.7). This suggests that the dna binding target is strongly conserved, just the opposite of PRDM7/9. However this conservation in ZNF133 weakens markedly in the distal 3 repeats.
The 11 conserved zinc fingers in ZNF133 are long enough to specify nearly unique dna sites in a 3 gbp genome, even if not all fingers take part in a given site recognition. Note the SGEKP lockdown cap departs from canonical form in repeats 5, 7, 12, and 13 perhaps impacting binding site utility. Human variation in repeat numbers has not been studied but it appears from phylogenetic considerations to be far less common than in PRDM7/9. Dotplots of ZNF133 show far less agreement across repeats at the dna level, indicating that neither homogenization, expansion, nor contraction of repeats by replication slippage has occurred recently in this gene (unlike PRDM9).
Alignment of human ZNF133 zinc finger array to orthologs in Primates, Glires, Laurasiatheres, Xenarthra and Afrotheres
z z z z z z z z z z z z z z z z z z z z
homSap VNCGECGLSFSKMTNLLSHQRIHSGEKP YVCGVCEKGFSLKKSLARHQKAHSGEKP IVCRECGRGFNRKSTLIIHERTHSGEKP YMCSECGRGFSQKSNLIIHQRTHSGEKP YVCRECGKGFSQKSAVVRHQRTHLEEKT
calJac ............................ ............................ ............................ ............................ ............................
oryCun ........G...LA.............. ............................ ............................ ...T........................ ............................
equCab ........G................... ............................ ............................ ............................ ............................
canFam ...R....G................... ............................ ............................ ...................R........ ............................
dasNov I..A....G................... ............................ ............................ ............................ .......................S....
proCap ...E....G................... ............................ ............................ ........R................... .......................S....
homSap IVCSDCGLGFSDRSNLISHQRTHSGEKP YACKECGRCFRQRTTLVNHQRTHSKEKP YVCGVCGHSFSQNSTLISHRRTHTGEKP YVCGVCGRGFSLKSHLNRHQNIHSGEKP IVCKDCGRGFSQQSNLIRHQRTHSGEKP
calJac ............................ ............................ ............................ ............................ ............................
oryCun ...G........................ ............................ ............................ .....................T...... ...Q......................R.
equCab ............................ ............................ ............................ .........................D.. ............................
canFam ...N........................ ............................ ............................ .........................D.. ............................
dasNov ............................ ............................ ............................ ................I........D.. ............................
proCap ............................ ...G........................ ............................ ................T........D.. ............................
homSap MVCGECGRGFSQKSNLVAHQRTHSGERP YVCRECGRGFSHQAGLIRHKRKHSREKP YMCRQCGLGFGNKSALITHKRAHSEEKP CVCRECGQGFLQKSHLTLHQMTHTGEKP YVCKTCGRGFSLKSHLSRHRKTTS VHHRLPVQPDPEPCAGQPSDSLYSL
calJac ...A......................K. ............................ ............................ ..........I............N.... ....M..Q..............K. ......L..G...R....A...C..
oryCun ...Q............L.........K. ............................ .T.........S.........T...... .GGGQ...S.S......S..L..K.... H......Q...Q.........IKA ...KP.LH..S.AYS...PGP....
equCab ...E............I.........K. ............................ .T........S.........W......L ..........I.....V......Q...L .......Q...Q........RMK. ..Q.P.PH.AS.A.S..S..P.H..
canFam ...E......................K. ............................ ..........S......I...V...... ........D.I.....L......Q.... ....M.DK...H........RMK. ..YK..LP....A....S..L.H..
dasNov ...E............I.........K. ...................R........ .T........S...T......L...... ........S.I.R...I....I.KE... ...R...Q...Q.......SRMKC ...KPLL...S.DYS..S..P....
proCap ...ED...........I.........K. ............................ .A....R...N...T..A..QL...D.L .......ED.M.....LV.....K.... ..SR.H.Q..NQ......Y.RIK. ...KS.F.S.L.T.S..S.VPV...
The ubiquitously expressed ZNF133 has been established by experiment to be a transcriptional repressor, recognizing specific sites in dsDNA. Despite the presence of the KRAB domain (which usually has this task), the zinc finger array alone contributes to transcriptional repression, with this effect mediated by another gene product, PIAS1, which binds the main array and recruits histone deacylases. The early zinc finger is not necessary for the PIAS1 effect and though conserved, its role remains obscure. PIAS1 may also have a role in PRDM9 and recombination.
For ZNF133, the weblogo below based on 413 repeats from 32 placentals illustrates that quite different selectional pressures have been operative here than in PRDM7/9. First, variation is not concentrated at the four special amino acid positions (purple boxes between CxxC HxxxH) but instead is distributed (though unevenly) among the non-C2H2 positions. Some of this occurs at residues primarily concerned with the zinc binding fold and not targeting macromolecule interactions. This establishes structural variation in the fold can be tolerated, ie PRDM7/9 is the real oddity for not exhibiting it.
The early zinc finger (which is classified by Pfam as C2H2)in the terminal exon is rather variable. While a consistently found zinc finger in such a protein is suggestive, nothing can be said about its function at this time.
early zinc finger of ZNF133 early zinc finger of PRDM7/9 early zinc finger of ZNF343 early zinc finger of ZNF589
homSap YLDPFCPPGFSSQKFPMQHVLCNHPPW HPCPSCCLAFSSQKFLSQHVERNHSSQ YTCSSCLLAFSCQQFLSQHVLQIFLGL YTCSSCLLAFSCQQFLSQHVLQIFLGL
panTro ........................... ........................... ........................... ....C.......P..............
gorGor ........................... ........................... ..........L................ ...........................
ponAbe ........................... ........................... ........................... ...........................
rheMac ........................... .........................T. .P......................... ...........................
papHam ........................... .........................T. .P......................... ...........................
calJac C.......................... .................H......... .P......................... .......VV..................
micMur H.G..F..DL......V.R...S.... ......S.............KHT.... .P.......S.........T....Q.. ..FWL......................
otoGar H.G.L...DL......R...P...... ......S....T..........T.P.. .P..................FR..... (no seqs before duplication)
tupBel H.SVS..LD...E......E....H.. ...L..S.........N....H...C. (no seqs before duplication)
cavPor Q.G..GG.D..A.R..V.....GQ... ......S.....H......M.CS....
oryCun H.G.L...DC.T..L.V..T..DP... ...FL.S.........T....W..RTE
ochPri S.G.C....L...N....QP.GDP.R. ...A..S.............QH..P..
turTru H.G..R..D....QLR...M..S.... Q..G..S.......I......CS.P..
bosTau H.C.....DLC....H..Q...SP... ......S......R........S.P..
equCab H.C.....D.....VH..R........ .R....S..............CK....
felCat H.C....SD..-L..H...M..T.... ......S............L.H..P..
canFam H.C.L..SD.....RHT..M....... ......SV.....T.....GK...P.E
myoLuc H.CA....D......H...M..SN... ......S.................P..
eriEur PSC.SN..DI....SH...MP...C.. Y...C.S....N.....R...HS.P.L
sorAra H.C....SD.....LHV.R........ ......R............MKHS.P.P
loxAfr QPC.....D......H..R...SP... N.....P..L...QLKHS.PFQSLPGT
proCap Q.C.....N..G...H...A....R.. ......P....TP....H..KHS.PC.
echTel QPC....LD..N...HK.....S.A.. ......P....TE.......Q...P..
dasNov QFC.....D...K..H......S.... ......P....T.....Y..NHS...E
HKR1 is another zinc finger protein that often surfaces in PRDM9-related blast searches. Structurally it is very similar to ZNF133. The zinc finger array begins with two very degenerate units that cannot bind zinc but may still retain the fold. The next 9 fingers are conventional but the tenth is missing the last two amino acids of the SGEKP cap. The last repeat has an intercalating residue after the two cysteines and lacks the final 3 cap residues. These features were in place at the time of stem placental divergence.
This gene, sometimes called ZNF875, also arose in placental mammal as part of the dramatic expansion of zinc finger proteins. However regulation of gene expression is probably no more refined in placentals than in marsupials, birds and other vertebrates -- these just have different systems. Indeed, rodents seem to have lost both HKR1 and ZNF133 yet get along just fine with poor overall orthologous correlation to primates.
The intra-repeat pattern of variation is different than in PRDM9 and ZNF133. There is more of it and this variation is not concentrated on the macromolecule recognizing amino acid positions, in fact seems to avoid it. This implies that the binding partner is fixed. The single 1998 publication on this gene sheds no light on what this might be. Assuming the function is in regulation of gene expression, the recognition sites in human might be predicted approximately from the conserved zinc fingers. This would yield an association with specific genes including false positives and negatives. Repeating this exercise in a dozen mammals and identifying the commonalities to the human gene set would yield a much improved list of regulated genes. HKR1 is widely expressed in a variety of tissues.
z z z z z z z z z z z z z
HKR1_homSap IKYEEFGPGFIKESNLLSLQKTQTGETP YMYTEWGDSFGSMSVLIKNPRTHSGGKP YVCRECGRGFTWKSNLITHQRTHSGEKP YVCKDCGRGFTWKSNLFTHQRTHSGLKP YVCKECGQSFSLKSNLITHQRAHTGEKP
HKR1_panTro2 ............................ ..............I............. .G.......................... ............................ ............................
HKR1_ponAbe2 ........D.......F.F......... ..............I.........R... ............................ ....H...............GI...... .M..........................
HKR1_papHam1 ...........................A ..............I............. ............................ ............................ ............................
HKR1_calJac1 ............K......R.......A .V.....Q.........G.......... R........................... ..........S...........R..... ....D...............K.......
HKR1_tarSyr1 ..C.........N....NF...H....A .......Q..S.V.......K....E.. .M.........................A .........................V.. F...........................
HKR1_micMur1 .......R....DP...GF...H....T .......Q..S..............E.. ...G........................ A.......................V... M..........................
HKR1_dipOrd1 L...KL..R.M.....P.....HPR..S FIG.K..Q.LSRLP..M...K..V.D.. FL.Q.................M...... F....................I...V.. .M.Q.................S......
HKR1_equCab2 ...R...L..............H...I. R..S...Q..SN....T..QSMR..E.. ...G............V........... ....E....................VR. .......................S....
HKR1_canFam2 .......L..L..PK......MGA.... ......KQ..SKR.I....QKIP..EN. ...K........................ ....E....................V.. .......................S....
HKR1_proCap1 .G.GDL.L...RG.D......AY..G.T .LCN...RDL........KQ..R.R... H..S............L........... H..AE...A.A.R..........A.... HG.RD.....R..A..AA.R...A.AR.
HKR1_dasNov2 ..CTD..F.C..K..V......NIA.SS ...S...EG.N...I....R..Q.EE.. ..........N................. ....E................I...V.. .I.....................S....
HKR1_choHof1 M.CG...L....K..V......HI...A ...S..ERG.S...I....Q....EE.. ..........N......A.......... ....E................I...V.. .I.....................S....
z z z z z z z z z z z z z z z z z z z z
HKR1_homSap YVCRECGRGFRQHSHLVRHKRTHSGEKP YICRECEQGFSQKSHLIRHLRTHTGEKP YVCTECGRHFSWKSNLKTHQRTHSGVKP YVCLECGQCFSLKSNLNKHQRSHTGEKP FVCTECGRGFTRKSTLSTHQRTHSGEKP
HKR1_panTro2 ......................................................... ............................ ............................ ................I...........
HKR1_ponAbe2 ............................ ............................ ............................ ............................ ................I...........
HKR1_papHam1 ............................ ............................ ............................ .A.......................... ...A............I...........
HKR1_calJac1 .......H........I..R........ .T.......................... ......W.Q................... ..F......................... ...MA.......R...I...........
HKR1_tarSyr1 ......E.........I..R.I...... .V....K..................... .....................M...... ........R..........R........ ................N...........
HKR1_micMur1 ................I........... .V......A................... ............................ ............................ ................I...........
HKR1_dipOrd1 ...K...S........I........... FV....Q.R.......V........... .I......G.......L........... ........S.......S...KA.A.... .G......S.....S.V...KK.....L
HKR1_equCab2 ................I........... .V.......................... ..........................R. .T......R.................... ..R............I...........
HKR1_canFam2 .......H........I........... .V....D.S................... .I.......................... ........R.................... ..R............I...........
HKR1_proCap1 H..A....A.G.S...A..A......R. HA.GQ...A.G.....V........... F........................I.. ......E................S..... ..Q.........T..V...........
HKR1_dasNov2 F......H....N...I..L........ .V.......................... ...P.....................I.. .M.....................S..... I.R............I...........
HKR1_choHof1 F......H....N...I..L........ .V.......................... .......................A.I.. ................S......S..... K.R...Q........IS..........
z z z z z z z z z z z z z z z z z z z z
HKR1_homSap FVCAECGRGFNDKSTLISHQRTHSGEKP FMCRECGRRFRQKPNLFRHKRAHSGA FVCRECGQGFCAKLTLIKHQRAHAGGKP HVCRECGQGFSRQSHLIRHQRTHSGEKP YICRKCGRGFSRKSNLIRHQRTHSG
HKR1_panTro2 ............................ .......................... ............................ ............................ .........................
HKR1_ponAbe2 ............................ .......................... .......................S.... ............................ .........................
HKR1_papHam1 ...R............V........... .......................... .......................S.... ..........N................. .........................
HKR1_calJac1 ...R........................ .T........................ ....G...A..D.....N.H.E.S...L ..............Y............. ...........W.............
HKR1_tarSyr1 ...R........................ .......................... ...G.......D....L...KE.SA... ...P....D...K............... .V......C.......V.......R
HKR1_micMur1 ...R........................ .V.......................S C.......S..D...F.......L.... ............................ ........C...........K....
HKR1_dipOrd1 ...R..K.S....L..HT...I...... .......Q.................D S..........D.......E...S.N.V .M..............L........... ......A.A.......L.....I..
HKR1_equCab2 ...V........................ .I........G............A.. ...........D.....T.....S...S ................A....I...... .T....................G..
HKR1_canFam2 ...R.............A.......... .I.....K..S........R.....T ...........D.....T..K..S.... .....................I...... ......E................P.
HKR1_proCap1 ...R....A.....A.L........... ...G......S.R......R.T.... L..K.......D....NA.....S..R. ...V......G.....V........... .V....E..................
HKR1_dasNov2 ...R........................ .I..Q.....S............... ...........D.....T.....S.... ............................ .........................
HKR1_choHof1 ...R........................ .I.K......S..............V ....D......D.....T.....S.S.. ......R..................... .V.......................
HRK1 is found within a cluster of ZNF genes on chromosome 19 but has no better than 50% identity to any of them. PRDM7, PRDM9, ZNF133, ZNF343, ZNF589, ZNF169 and ZNF596 are not found in tandem ZNF clusters nor in syntenic associations, as determined by setting the UCSC GeneSorter tool to gene distance and comparing gene neighbors.
ZNF343 and ZNF589
ZNF343 is another very closely related KRAB zinc finger array protein. It appears restricted phylogenetically to primates but may have earlier spin-offs reminiscent of PRDM7 (such as the microbat sequence below). Two species, baboon and tarsier have deletions of 2 and 1 zinc finger respectively. The new world monkey Callithrix has a moderately degenerated pseudogene. However it is not plausible that other mammalian species (such as rodents) ever had this gene.
No experimental study has ever considered the function of this gene though occasionally it surfaces in expression studies. It is quite conserved at least in apes, indicative of an important function in gene regulation via specific site recognition. Outside of the terminal zinc finger region, ZNF589 most closely resembles ZNF589 and ZNF133 and so has no direct bearing on PRDM7 or PrDM9.
Prior to ZNF gene family expansion, each of the proteins initially present may have had multiple functions (as proposed by Piatigorsky). With zinc finger arrays, different subsets of fingers may have recognized different dna sites, regulating different genes. After gene duplication, the descendent genes could then diverge to specialize on distinct subsets of these pre-existing functions (subfunctionalization). This allowed fine-tuning of regulation relative to a parent gene operating under slightly conflicting selectional pressures that had to be satisfied simultaneously.
Placental mammals today do not differ greatly from the stem placental mammal in which the expansion began. This expansion (and later contraction) continued into the present era, yielding many lineage-specific sets of ZNF gene families (ie lack of 1:1 orthologous correspondences). Evolution need not take the same path to reach the same end -- indeed, marsupials, birds and other vertebrates have attained excellent regulation of gene expression by other approaches.
ZNF343_homSap INCREYEPDHNLESNFITNPRTLLGKKP YICSDCGRSFKDRSTLIRHHRIHSMEKP YVCSECGRGFSQKSNLSRHQRTHSEEKP YLCRECGQSFRSKSILNRHQWTHSEEKP YVCSECGRGFSEKSSFIRHQRTHSGEKP ZNF343_panTro ...................S........ ............................ ............................ ................S........... ............................ ZNF343_gorGor ............................ ............................ ............................ ............................ ............................ ZNF343_ponAbe .....C...................... ..............A............. ............................ ............................ ............................ ZNF343_nomLeu .....C...............I...... .....................T...... ............................ ...........N................ ............................ ZNF343_macMul .....C...RS................. ..............A......T...... ............................ ...........N....K........... ............................ ZNF343_papHam .....C...RS................. ..............A......T...... ............................ ...........NN...K........... ....D....................... ZNF343_tarSyr N..K.R...YSP.....R.S..F..E.. CV......G..N....N..R.T..V... ....D.....KNR.T.I.......G... .VR.Q..RG.SQ..NVAQ..R...D... .I.R......RD..TLVI.E........ ZNF343_otoGar V....F.S.C...........V.FRE.. .V.....PG.....I......T.TG... .E..............T..R........ ...........N....S.......G... .M....E....Q................ ZNF343_myoLuc GS.N.H.L.CS.K...AV.QV..SEE.. .V.RE...G.NNK.N.N....T...... ...GD......LMAI.VH......G... .V.K...RG.SK..N....TE....... .L.R...QS.RNN.VL....WI...... ZNF343_homSap YVCLECGRSFCDKSTLRKHQRIHSGEKP YVCRECGRGFSQNSDLIKHQRTHLDEKP YVCRECGRGFCDKSTLIIHERTHSGEKP YVCGECGRGFSRKSLLLVHQRTHSGEKH YVCRECRRGFSQKSNLIRHQRTHSNEKP ZNF343_panTro ............................ ............................ ............................ ............................ ............................ ZNF343_gorGor ............................ ............................ ............................ ............................ ............................ ZNF343_ponAbe ........G................... ...K........................ ...G........................ ............................ ............................ ZNF343_nomLeu ........G................... ............................ ............................ ............................ ............................ ZNF343_macMul ........G................... ............................ ............................ ............T............... ............................ ZNF343_papHam ---------------------------- ---------------------------- .......G.................... ............T............... ............................ ZNF343_tarSyr F..S.Y.QG.IQ..Q.LV...T....N. ---------------------------- ....K.....SW..H.LV.Q.K...... ...R....S..Q..CVIT.........P .I....G....K..S.........G... ZNF343_otoGar .I......G.S..........T...D.. ......R.....K.N..R.....SN... .I............N..V...M...... .T.S........................ ......G..Y.............A.... ZNF343_myoLuc ...P....G.AY.........T...... .I.Q...H...EK.SF.R.....SG... F..L......G.....RK.Q........ .T.S....S.TQ..F..I..G....... ......G.S..Y........K...DV.. ZNF343_homSap YICRECGRGFCDKSTLIVHERTHSGEKP YVCSECGRGFSRKSLLLVHQRTHSGEKH YVCRECGRGFSHKSNLIRHQRTH ZNF343_panTro ............................ ............................ ....................... ZNF343_gorGor ............................ ............................ ..........G............ ZNF343_ponAbe F........................... ................P........... ....................... ZNF343_nomLeu ..............S............. ...................K........ ....................... ZNF343_macMul ............................ .....................I...... ................V...... ZNF343_papHam ............................ .....................I...... ....................... ZNF343_tarSyr .V.K......SQ..Y..K.Q...LD... FI.R.......W...............P ...........Q..Y..K.E... ZNF343_otoGar .V.G..........A............. .IR.DR...S.Q....VS.........W ..........GY........... ZNF343_myoLuc ..........FY..D..I.......... ........S..Q..F.VI..G....K.. ....D...S..YR....T...K.
ZNF589 is also a primate-specific expansion within the KRAB ZNF gene family which may have expanded independently from the same parental gene in artiodactyls, again similarly to the separate expansions of PRDM7. It has been the subject of 3 publications under the name SZF1 and its consensus region identified experimentally as CCAGGGTAACAGCCG which is similar to that of ZBRK1. Regulation of gene expression reportedly takes place in hematopoietic progenitor cells.
In humans, ZNF589 has an internal stop codon at the second cysteine within the 5th repeat due to a T to A transversion. This is not an error or mutation in the reference genome hg19, nor a balanced polymorphism, nor 1% allele as no corrective SNP is known from the 1000 genome project or individual sequencing projects. It remains possible that some human populations (notably African because of greater diversity) will not have this stop codon if it is a very recent development. However all human populations sampled including bushman KB1 genomic reads all contain it, as do Neanderthal and Denisova fossil dna. (View chr3:48,285,273-48,285,286 on UCSC human genome browser hg19 with appropriate tracks opened.)
Past the stop codon, the zinc finger array continues on another nine repeats and do not seem impaired (strict conservation of cysteines, histidines, and invariant phenylalanine and leucine). It is not clear whether the mRNA would be targeted by nonsense mediated decay, whether a truncated and possibly still functional protein is produced, or whether that a suppressor mechanism that allows some read-through of the early stop codon. If ZNF589 functions with four repeats, the dna recognition sites would be truncated relative to what the full zinc finger array could have recognized. Terminal alternative splicing has been seen for ZNF589 but not rejoin the array.
Chimpanzee does not have this internal stop codon and has a full set of repeats. However the orthologous gene in gorilla (contig CABD02243014) has a frameshift near the end of its 10th repeat. Assuming this is not assembly error in a low coverage genome, this raises the same question about pseudogenization vs function-retaining truncation as in human. If this represents gene loss, the event is independent from the one in human and the resultant protein may have higher dna specificity (recognize a longer or different site).
Similarly orangutan has a frameshift at the end of the 7th repeat, a reading frame restoring frameshift 4 repeats later followed shortly by an early stop codon. Gibbon also has a frameshift. However macaque, like chimpanzee, has maintained a full length zinc finger array. New world monkey Callithrix has a much older pseudogene. This is truly an ortholog because biflanking synteny is still preserved (NME6+ ZNF589- CAMP-) from human to marmoset.
Tarsier also has severely decayed gene candidate. Its contig maps uniquely to the ZNF589 region of human by blastn but is too short to establish flanking synteny. Since ZNF589 appears missing in lemurs and tree shrew, the duplication event occurred in the tarsier divergence stem, or about 65 myr ago. Tarsiers are basal haplorrhine and not part of the strepsirrhine (lemur) clade.
Thus the gene duplicate ZNF589 persisted in the ancestor past the tarsier and new world monkey divergences and losses, kept a full length repeat role in old world monkeys and chimp, but may have carved out altered separate roles in great apes with utilizing recently truncated arrays.
The parental gene for ZNF589 -- based on the terminal exon minus the array -- appears to be ZNF133 or PRDM7, genes that arose about the same time in stem placentals.
ZNF589 Repeat Region in Apes: nominal length of zinc finger array produced shown in blue stop codons and truncated repeats shown in red frameshifts and cryptic repeats shown in purple
>ZNF589_homSap AADD01032563 QVCRECGRGFSRKSQLIIHQRTHTGEKP YVCGECGRGFIVESVLRNHLSTHSGEKP YVCSHCGRGFSCKPYLIRHQRTHTREKS FMCTVCGRGFREKSELIKHQRIHTGDKP YVCRD*GRGFVRRSCLNTHQRIHSDEKP FVCRECGRGFRAKSTLLLHQWTHSEVKP HVCEECGHGFSQKSSLKSHRRTHSGEKP YVCGECGRGFSRRIVLNGHWRTHTGEKP YTCFECGRNFSLKSALSVHQRIHSGEKP YACTECGQGFITKSQLIRHQRTHTGEKP YVCGECGRGFIAQSTLHYHRSTHSKEKP YVCSQCGRGFCDKSTLLAHEQTHSGEKP YVCGECGRGFGRKILLNRHWRTHTGEKP YACIECGRNFSHKSTLSLHQRIHSGEKP YACVECGQSFRRKSQLIIHQKIHSGKSF RGARSEDVILATSQPSATPAEMLREKPCL |
>ZNF589_panTro AADA01029841
QVCRECGRGFSRKSQLIIHQRTHTGEKP
YVCGECGRGFIVESVLRNHLSTHSGEKP
YVCSHCGRGFSCKPYLIRHQRTHTREKS
FMCTVCGRGFREKSELIKHQRIHTGDKP
YVCRDCGRGFVRRSCLNAHQRIHSDEKP
FVCRECGRGFRAKSTLLLHQWTHSEVKP
HVCEECGHGFSQKSSLKSHRRTHSGEKP
YVCGECGRGFSRRIVLNGHWRTHTGEKP
YTCFECGRNFSLKSALSVHQRIHSGEKP
YACTECGQGFITKSQLIRHQRTHTGEKP
YVCGECGRGFIAQSTLHYHRSTHSKEKP
YVCSQCGRGFCDKSTLLAHEQTHSGEKP
YVCGECGRGFGRKILLNRHWRTHTGEKP
YACIECGRNFSHKSTLSLHQRIHSGEKP
YACVECGRSFRRKSQLIIHQKIHSGKSF
RGARSEDVILATSQPSATPAEMLREKPCL
|
>ZNF589_gorGor CABD02243014 QVCRDCGRGFSRKSQLIIHQRTHTGEKP YVCGECGRGFIVESVLRNHLSTHSGEKP YVCSHCGRGFSCKPYLIRHQRTHTREKS FMCTVCGRGFREKSELIKHQRIHTGDKP YVCRDCGRGFVRRSCLNTHQRIHSDEKP FVCRECGRGFRAKSTLLLHQWTHSEVKP HVCEECGHGFSQKSSLKSHRRTHSGEKP YVCGECGRGFSRRIVLNGHWRTHTGEKP YTCFECGRNFSLKSALSVHQRIHSGEKP YACTECGQGFITKSQLIRHQRTHTgEKP YVCGECGRGFIAQSTLHYHRSTHSKEKP YVCSQCGRGFCDKSTLLAHERTHSGEKP YVCGECGRGFGRKILLNRHWRTHTGEKP YACIECGRNFSHKSTLSLHQRIHSGEKP YACMECGRGFRRKSQLIIHQKIHSGKSF RGARSEDVILATSQPSATPAEMLREKTCL |
>ZNF589_ponAbe ABGA01071880 QVCRECGRGFSRKSQLIIHQRTHTGEKP YVCRECGRGFIVESVLRNHLSTHSGEKP YVCSHCGRGFSCKPYLIRHQRTHTREKS FMCTVCGQGFREKSELIKHQRIHTGDKP YVCRDCGRGFVRRSCLNTHQRIHSDEKP FVCKECGRGFHAKSTLLLHQWTHSEVKP HVCEECGHGFSQKSTLKSHRRTHSGeKS YVCEECGRGFSRRIFLNGHWRTHTREKP YTCFECGRNFSLKSALSVHQRMHSGEKP YACTECGQGFITKSQLIRHQRTHTGEKP YVCREWARLYSSDNPPLPPAYTLQGETp YVCSQRG*GFCDKSTLLAHEQTHSGEKP YVCGECGWGFGRKILLNRHWRTHTGEKT YACIECGQNFSHKSTLSLHQRIHSGEKP YACMECGRGFRRKSQLIIHQKIHSGKSF RGASSEDVILATSQPSATPAEMLREKTCL |
>ZNF589_nomLeu ADFV01172942 QVCRECGRGFSRKSQLIIQQRTHTGEK YVCEECGRGFIVESVLRNHLSAHSAEKP YVCSHCGRGFSCKPYLMRHQRTHTREKS FMCTVCGRGFREKSELIKHQIIHTGGKP YVCRDCGRGFVRRSCLNTHQRIHSDEKP FVCRECGRGFRAKSTLLLHQWTHSEVKP HVCEDCGHGFSQKSTLKSHRRTHSGEKP YVCGECGQGFSRRIFLNGHWRTYTGEKP YTCFECGRNFSLKSALSVHQRIYWGEkP YACVECGRGFITKSQLIRHQRTHTGEKP YVCGECGQGFIAQSALRYHRSTHSREKP YVCSQCGrGEAFVINQLAHEQTHSGEKP YVCGECGQGFGRKILLNRHWRTHTGEKP YACIECGRNFSHKSTLSLHQRIHSGEKP YACTECGRGFRRKSQLITHQKTHSGKSF RGARSEDVILATSQPSATLAEMLREKACL |
>ZNF589_rheMac AANU01238696
QVCGECGRGFSRKSQLIIHQRTHTGEKP
YVCGECGRGFIVESVLRNHLSTHSGEKP
YVCSQCGRGFSCKPYLIRHQRTHTREKS
FMCTVCGRGFREKSELIKHQRIHTGDKP
YVCRDCGRGFVRRSCLNTHQRIHSDEKP
FVCRECGRGFRAKSTLLLHQWTHSEVKP
HVCEECGHGFSQKSTLKSHQRTHSGEKP
YVCGECGRGFSRRIFLSGHWRTHTGEKP
YTCFECGRNFSLKSALSVHQRIHSGEKP
YACAECGRGFITKSQLIRHQRTHTGEKP
YVCGECGRGFIAQSTLHYHRSTHSGEKP
YVCSQCGRGFRDKSALLAHEQTHSGEKP
YVCGECGWGFGRKILLSRHWRTHTGEKP
YACMECGRNFSHKSTLSLHQRIHSGEKP
YACTECGRGFRRKSQLSIHQKTHLGKSF
RGARSEDVIFASQPSAAPAEMLREKPCL
|
>ZNF589_papHam ti|2005908815
QVCRECGRGFSRKSQLIIHQRTHTGEKP
YVCGECGRGFIVESVLRNHLSTHSGEKP
YVCSQCGRGFICKPYLIRHQRTHTREKS
FMCTVCGRGFRENSELIKHQRIHTGDKP
YVCRDCGRGFVRRSYLNTHQRIHSDEKP
FVCRECGRGFRAKSTLLLHQWTHSEVKP
HVCEECGHGFSQKSTLKSHRRTHSGEKP
YVCGECGRGFSRRIFLSGHWRTHTGEKP
YTCFECGRNFSLKSALSVHQRIHSGEKP
YACAECGRGFITKSQLIRHQRTHTGEKP
YVCGECGRGFIAQSTLHYHRSTHSGEKP
YVCSQCGRGFRDKSTLLAHEQTHSGEKP
YVCGECGRGFGRKILLSRHWRTHTGEKP
YACMECGRNFSHKSTLSLHQRIHSGEKP
YACTECGRGFRRKSQLNIHQKTHLGKSF
RGARSEDVIFASQPSAAPAEMLREKPCL
|
>ZNF589_calJac ACFV01038884 QMCTVCGRGIRNKSHLIQHQRIHTGDKP YVCRNCGRGFVRSCLIK HQRILSGEKP FICRECGRGFRDKSTPHThQRAHSGEKP sCGEECGRGFTRKSTLKSHRRTHSGEKP YVYGECGWGFSSKGVLNTHWRTHTGAKP YACRVATSPLsHKSTLSSHQRIHSGEKP |
>ZNF589_tarSyr ABRT010411760 SVCREYGQSFSRKSHLLRHWRTHTGEKP YVnGNCGHTFIDKSVLHNYQSTHSGKKP YVCRECGCSLD*KSHLIRHQRTHTQERP FMCTV |
By comparing each sequence to itself and to PRDM9, it emerges that PRDM9 is highly unusual for its remarkable self-similarity in its zinc finger array. That strongly suggests some form of homogenization (master-slave) of repeats that is unique to it and very likely highly relevant to its role in defining recombination hotspots. Whatever the precise evolutionary relationships to the other closely related zinc fingers, that has not resulted in retention of close matching at the dna level to PRDM9.
The alignment below of the final exon (minus the terminal arrays) shows positions conserved at 60% identity. Overall, conservation is unremarkable outside the early zinc finger region. However several distal patches also suggest a moderate level of conservation pressure and thus some function beyond merely serving as a long linker between the terminal array (which is wrapped around the major groove of the dna target) and the early repeat (whose binding partner, if any, is unknown).
first anomalous repeat PRDM9_homSap E.KPEI..CPSC.LAFSSQ.FLSQHV..NH..Q.F....A...L.P.NP.PGDQ.Q.-QQ..D.........GQE....S..L..RT..R....AFSSPP.-.Q..S.R.G.R..E.E....Q..NP..T.K............ ....ECG.GFS..S....HQRTHTGEK. PRDM7_homSap E.KPEI..CPSC.LAFSSQ.FLSQHV..NH..Q.F....A...L.P.NP.PGDQ.Q.-.Q..D.........GQE....S..L..RT..R....AFSSPP.-.Q..SSR.G.R..E.E....Q..NP..T.K............ ....ECG.GFS..S....HQRTHTG.KP PRDM7_calJac E.KPEI..CPSC.LAFSSQ.FLS.HV..NH..Q.F........L.P.NP.PG.Q...-QQ..D.........GQE....S..L..RT..R....AFS.PP.-.Q..SSR...R..E.E....Q..NP..T.K............ ....ECG.GFS..S....HQRTHTGEKP ZNF133_homSap ...PE....P.C...FSSQ.F..QHV..NH....F....A.....P..P.PGDQ...-QQ............G.E.....-.L..RT..R-...AFS.PP.-.Q..SSR.G.R..E.E....Q..NP..T.K............ ....ECG..FS.......HQR.H.GEKP ZNF589_homSap E.KPE...CPSC.LAF.SQ.FLSQ....NH....F.---A...L.P.NP.P.DQ.Q.-Q...D..........Q........L...T..R.....FSS...-....SS..G.R..E......Q..................... ....ECG.GFS..S....HQRTHTGEK ZNF343_homSap E.KPEI..C.SC.LAFS.Q.FLSQHV.-----Q.F....A.....P.N..PG...Q..QQ............GQE....S.....RT..R....AF.SP..-.Q..S.R.G....E.E....Q..NP....K............ ....E........S......RT..G.KP HKR1_homSap E.KPEI...PSC.L.FSSQ..LSQHV...H..Q.F....A...L......P.DQ.Q.----.D................S..L..R........A.SSPP...Q...S.....................T.K............ ....E.G.GF...S.....Q.T.TGE.P
Domain by domain structure/function
PRDM7 and PRDM9 are chimeric proteins comprised of 6 recognizable domains joined by linker regions. While multi-domain proteins are common in the overall human proteome, this particular combination occurs nowhere else. However some of the domains here occur in other combinations in other proteins, notable in the vast heterogeneous family of zinc finger proteins (gene names ZNFxxx).
KRAB_A Kruppel SSXRD zinc knuckle PR or SET domain early zinc finger terminal zinc finger array
Because the inter-domain linkers are evolving chaotically in terms of little amino acid property conservation and sometimes length, they cannot plausibly be under significant selective pressure, nor can they assume a stable structural fold. However this does not imply that the domains that they link do not have significant physical interactions important to the global tertiary protein structure. To date, only the isolated domains have been studied crystallographically (with the exception of the knuckle-PR combination).
While the domain folds individually are quite ancient and do not reflect de novo innovation in vertebrates from random dna strings, their assembly into PRDM7/9 is fairly recent, about 150 million years ago. Prior to this, a proto-PRDM7 containing the last 4 domains arose and persisted for 300 million years, giving rise to several gene duplicates, all with vaguely understood function related to transcriptional regulation.
The following sections consider what is known about each domain in turn primarily from the perspective of comparative genomics. As of July 2011, 51 land vertebrate genomes are available, providing a rich history of how PRDM7 has been evolving in various branches of the phylogenetic tree.
Reciprocal translocation: origin of the SSX1-PRDM chimera
Upon blastp of the first 6 exons of any PRDM7/9 protein against GenBank restricted to human, SSX1 emerges as the only full length non-self match. Comparison of its 6 exons establishes further that their intron phasing is an exact match. Since this is impossibly coincidental, it follows that PRDM7 (the immediate parent of PRDM9 in primates) arose as a chimera of ancestors to these two proteins prior to marsupial divergence. The percent identity has dropped from the initial perfect agreement to 32% today, without however loss of KRAB_A and SSXRD domain recognizability in either gene family. No other proteins in the human genome -- in particular no zinc finger proteins -- contain these 6 exons though the KRAB domain alone is widespread.
>SSX1_homSap >PRDM9_homSap 0 MNGDDTFAKRPRDDAKASEKRSK 0 0 MSPEKSQEESPEEDTERTERKPM 0 0 AFDDIATYFSKKEWKKMKYSEKISYVYMKRNYKAMTKL 1 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GFKVTLPPFMCNKQATDFQGNDFDNDHNRRIQ 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VEHPQMTFGRLHRIIPK 0 2 VKPPWMALRVEQRKHQK 0 0 IMPKKPAEDENDSKGVSEASGPQNDGKQLHPPGKANISEKINKRS 1 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 2 GPKRGKHAWTHRLRERKQLVIYEEISDPEEDDE* 2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1 PRDM9 MSPEKSQEESPEEDTERTERKPMVKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITIGLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ M+ + + + P +D + +E++ K AF DI+ YF+K+EW +M EK Y +KRNY A+ +G + T P FMC+ +QA Q +D D + R Q SSX1 MNGDDTFAKRPRDDAKASEKRSK---AFDDIATYFSKKEWKKMKYSEKISYVYMKRNYKAMTKLGFKVTLPPFMCN-KQATDFQGNDF---DNDHNRRIQ PRDM9 VKPPWMALRVEQRKHQKGMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKLELRKKETERKMYSLRERKGHA-YKEVSEPQDDDYL 1 V+ P M R K MPK +E+ K +S ASG + K + P G+A+ S + ++ ++ + LRERK Y+E+S+P++DD SSX1 VEHPQMTFGRLHRIIPKIMPKKPAEDENDSKGVSE------ASGPQNDGKQLHPPGKANISEKINK-RSGPKRGKHAW-THRLRERKQLVIYEEISDPEEDDE*
This chimera arose subsequent to the duplication of proto-PRMD7 and its divergence to PRDM11, its nearest PRDM relative which has leading exons unrelated to SSX1. Indeed none of the other 14 PRDM proteins have a KRAB or SSXRD domain. The SSX1 gene itself, then and now, lies in a tandem array and so did not disappear as a standalone gene family as only one copy was used up in forming the hybrid protein. For viability, the event was likely a reciprocal translocation, accounting for the SSX array and PRDM7 being on different chromosomes today.
The SSX1-group genes occurs in the human reference genome as 11 features in two nearby clusters both on chromosome X. Some of these may be pseudogenes. The degree of similarity suggests recent gene duplication and/or gene conversion. The array is notorious for reciprocal translocations involving the one of 24 human synaptotagmins, the SYT4 gene on chromosome 18. These translocations fuse early exons of SYT4 with distal exons of an SSX gene, usually SSX1 or SSX2 but sometimes SSX4. The event takes place within intron 4 of the SSX genes and preserves reading frame, allowing for a chimeric protein with disastrous regulatory properties to emerge -- nearly all cases of synovial sarcomas arise from repeated occurrence of this event.
SSX1b + chrX:47967088-47980069 similar to SSX1 SSX5 - chrX:48045656-48056199 synovial sarcoma X breakpoint 5 SSX1a + chrX:48114797-48126879 synovial sarcoma X breakpoint 1 SSX9 - chrX:48154885-48165614 synovial sarcoma X breakpoint 9 SSX3 - chrX:48205863-48216142 synovial sarcoma X breakpoint 3 SSX4 + chrX:48242968-48252785 synovial sarcoma X breakpoint 4 SSX4B - chrX:48261524-48271344 synovial sarcoma X breakpoint 4B SSX8 + chrX:52651985-52662998 similar to SSX8 SSX7 - chrX:52673111-52683950 synovial sarcoma X breakpoint 7 SSX2a - chrX:52725946-52736249 synovial sarcoma X breakpoint 2 SSX2b + chrX:52780308-52790617 synovial sarcoma X breakpoint 2
Possibly the SSX1 array has long been predisposed to translocation events. It might seem very difficult to establish the structure of the ancestral array at the time of PRDM chimera formation -- contemporary marsupial has barely related genes on different chromosomes; elephant and dog too lack a multi-gene array. However rhesus but not marmoset has a chr X cluster, so that aspect is restricted to old world primates. A single SSX1 gene can be recovered from elephant but is already quite diverged from human. Marsupials have no evident SSX1 genes today.
This gene fusion of SSX1 and PRDM brought together a negative regulatory domain for transcription with a histone methylase and dna site recognition domain. This new combination succeeded in replacing whatever prior mechanism existed for meiotic breakpoint pairing and recombination.
>SSX1_loxAfr 0 VNRDSSLAKSSKEDTQKPEKESK 0 0 AFKDILKYFSKEEWAKLGYSKKVTYVYMKRNYDTMTNL 1 2 GLRATLPPFMDPNRLATKSQLDESDEEQNPGTQ 1 2 DEPPQMASSVRESKHLM 0 0 MKPKKPSKEENGSKVVPGTAGLMRTSGPEQAQKQPCPPGKANTSGQQSKQTP 1 2 VPGKEETKVWACRLRERKNLVAYEEISDPEEED*
The zinc knuckle preceding the PR (SET) domain
A 2011 crystallographic study establishes that a short motif YC..C..........C..HGP found in 6 members of the human PRDM gene family binds zinc via the 3 cysteines and a histidine. The fold most closely resembles the previously known RanBP2 zinc finger domain which occurs in some 21 human proteins, notably nucleoporins NUP153, NUP358, NPL4, EWS, TLS, RBP56, RBM5, RBM10, TEX13A, RANDB2 and ZRANB2. Not all these domains are necessarily homologous because the fold is small and zinc fingers seem to have evolved numerous times. Such fingers can bind other proteins, ssRNA and likely DNA. Their function in PRDM genes is completely unknown but the aromatic residue preceding the first cysteine may contribute to a pi-bonding base stack with guanines.
The domain begins at a phase 2 exon, meaning that the first codon letter is borrowed from the preceding exon splice donor. A dozen earlier residues from this exon are also used but do not exhibit any conservation outside their orthology class. In most cases the knuckle domain exon also contains a downstream PR(SET) domain but at variable intervening lengths (distances shown are to conserved FGP in center of PR(SET) domain. The function of these intervening residues are unknown.
exon 6 splice exon 7 SET gene name IPLNQHTSDPNN 1 2 RCDMCADNRNGECPMHGPLHSLRRLVG .49. PRDM6_homSap PDPPRPFDPHDL 1 2 WCEECNNAHASVCPKHGPLHPIPNRPV .16. PRDM10_homSap MAEDGSEEIMFI 1 2 WCEDCSQYHDSECPELGPVVMVKDSFV .99. PRDM15_homSap GSKENMATLFTI 1 2 WCTLCDRAYPSDCPEHGPVTFVPDTPI .36. PRDM4_homSap IVPKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPV .42. PRDM11_homSap KEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAV .42. PRDM9_homSap KEISEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAV .42. PRDM7_homSap QEIWDPQDDDYL 1 2 YCEECQTFFLETCAVHGPPKFVQDSVM .42. PRDM7_monDom NENYRPEDDDYL 1 2 YCEICQTFFLEKCVLHGPPVFVQDLPV .42. PRDM7_ornAna EEQDDTFNDQPF 1 2 YCEMCQQHFIDQCETHGPPSFTCDSPA .42. PRDM7_danRer TEEEELRDEEYF 1 2 FCEECKSFFIEECELHGPPLFIPDTPA .42. PRDM7_salSal IKEEEADVKDFL 1 2 YCEVCKSVFFSKCEVHGPALFIADSPV .42. PRDM7_ictPun YVCRECGRGFSWQSVLLTHQRTHTGEKP comparison to longer zinc finger in main array of PRDM7/9
Structural alignment of all PRDM proteins
To determine the evolutionary relationship of the 16 human PRDM genes, it is useful (given the great divergence in primary sequence) to consider rare genomic events such as intron gain/loss and indels. Only 7 of the 16 contain the knuckle region. Of these PDRM11 is the most closely related to PRMD9.
This is fortunate because the 3D structure of PRDM11 was recently determined (PDB: 3RAY) from before the knuckle region on into the final exon, thus allowing threading of PRDM9 (whose structure has not been studied). The dozen-odd conserved patches in these widely diverged paralogs find their explanation in the atomic details of this structure. Note the PRA(SET) domain and zinc fingers are all that can currently modeled as the KRAB, SSXRD and final exon have no counterparts at PDB.
The knuckle region apparently represents a one-time domain acquisition relative to a knuckle-less ancestral state. The date of this event relative to species phylogeny and the source of the domain are unclear (it is very unlikely to have evolved in situ). Similarly, the internal phase 00 intron is ancestral even though it breaks up a coherent structural domain. Note the final 12 intron is also ancestral -- the PR(SET) domain never occurs without it even though zinc fingers are not always found in the next exon. However the later 21 intron is a newer acquired feature specific to PRDM9 and its closest associates, post-dating acquisition of the knuckle domain and predating duplication and divergence of the PRDM7/9 group. This again follows from gene tree and parsimony considerations.
Crystallographic coverage is excessive yet highly unsatisfactory -- the knuckle-PR(SET) domain is covered by 6 different structures, yet none of them are exactly what is needed (PRDM11 3RAY; PRDM4 2L9Z/3DB5; PRDM10 3IXH; PRDM1 3DAL; PRDM2 3JV0; PRDM12 3EP0). There is no coverage of the preceding KRAB or SSXRD domain or the following early knuckle. However on the knuckle-PR(SET) domain, all these structures could likely be superimposed simultaneously on the near-universal domains identified below. PRDM7/9 would then follow this fold trace as well, though it could be modeled directly from just PRDM11. The intervening regions between conserved anchors can be modeled for PRDM7/9 only to the extent that local conservation in length and residue can be found to a determined structure. For example, IFYRTCRVI in PRDM9 can be modeled by the PRDM11 structure since its internal residues contain three matches and no gaps, IFYRACRDI.
Humans have 51 genes encoding SET domains, with the PRDM group most diverged from the canonical structures. It is difficult enough to meaningfully align the PRDM and even more so to include all 51 of these lysine methylases. When that is done, most of the conserved patches below emerge as universal motifs yet others are restricted to the PRDM family. All of these proteins would likely bind S-adenosyl methionine and have a lysine pocket in addition superimposable global folds (neither relatable to the 45 human arginine methyltransferases).
gapping: uncertain between conserved markers iM: initial methionine, protein thus too short for further comparison knuckle: shortened zinc finger motif underlining: magenta coloring shows non-informative idiosyncratic introns C2H2: terminal zinc finger region following universal phase 12 intron PRDM15: duplicated diverged exon removed 21 SWPASGHVHTQAGQGMRGYEDRDRADPQQLPEAVPAGLVRRLSGQQLPCRSTLTWGRLCHLVAQGR 0: indel unifying PRDM9/7/11, cannot be resolved as insertion or deletion 7: near-universal motif NWMrYV split by phase 21 intron gained by PRDM9/7/11/4 1: arginine supporting PRDM6 as outgroup to the knuckle subgroup 8: inexplicable repositioning of 6 residues to previous exon in PRDM4 2: near-universal motif SLP 9: near-universal motif EQNL 3: near-universal motif GF 10: near-universal motif IFY 4: indel unifying PRDM9/7/11, resolvable as an insertion 11: near-universal motif ELLVWY 5: near-universal motif FGP 12: possible synapormorphy grouping first 9 genes 6: near-universal motif WLI split by universal phase 00 intron PRDM16: CVDANQAGAG insertion removed from ISEDLGSEKFCVDANQAGAGSWLKYIRVA PRDM3: inexplicably has official gene name MECOM text-pdf version here
Applicable 3D structural determinations: .... PRDM9 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGI.PQAGLGVWNEASDLPLGLHFGPYEGRIT.....EDEEAANNGYSWLITKG.RNCYEYVD.......GKDKSWANWMRYVNCARDDEEQNLVAFQYHR..QIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 3RAY PRDM11 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTS...GESDVRCVNEVIPKGHIFGPYEGQIS......TQDKSAGFFSWLIVDK.NNRYKSID.......GSDETKANWMRYVVISREEREQNLLAFQHSE..RIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLAR 2L9Z PRDM4 WCTLCDRAYPSDCPEHGPVTFVPDTPIE....SRARLSLPKQLVLRQSIV..GAEVGVWTG.ETIPVRTCFGPLIGQQSHSMEVAEWTDKAVNHIWKIYHN.GVLEFCII.......TTDENECNWMMFVRKARNREEQNLVAYPHDG..KIFFCTSQDIPPENELLFYYSRDYAQQI.............. 3IXH PRDM10 WCEECNNAHASVCPKHGPLHPIPNRPVL....TRARASLPLVLYIDRFLG......GVFSK.RRIPKRTQFGPVEGPLV.....RGSELKDCYIHLKVSLDKGDRKERDLHEDLWFELSDETLCNWMMFVRPAQNHLEQNLVAYQYGH..HVYYTTIKNVEPKQELKVWYAASYAEFVNQKIHDISEEERK. 3DAL PRDM1 DGGTSVQAEASLPRNLLFKYATN.SEEVIGVMSK.EYIPKGTRFGPLIGEIY..TNDTVPKNANRKYFWRIYSR.GELHHFID.......GFNEEKSNWMRYVNPAHSPREQNLAACQNGM..NIYFYTIKPIPANQELLVWYCRDFAERLHYPYPGELTMMNL. 3JV0 PRDM2 LAEVPEHVLRGLPEEVR.LFPSAVDKTRIGVWAT.KPILKGKKFGPFVGDKK.....KRSQVKNNVYMWEVYYP.NLGWMCID.......ATDPEKGNWLRYVNWACSGEEQNLFPLEINR..AIYYKTLKPIAPGEELLVWYNGEDNPEIAAAIEEERASARSK 3EP0 PRDM12 SGEVQKLSSLVLPAEVIIAQSSIPGEGL.GIFSK.TWIKAGTEMGPFTGRVI..APEHVDICKNNNLMWEVFNEDGTVRYFID.......ASQEDHRSWMTYIKCARNEQEQNLEV.VQIGT.SIFYKAIEMIPPDQELLVWYGNSHNTFLGIPGVPGLEEDQKK
Phylogenetic variability of the knuckle-PR(SET) domain for PRDM7/9, shown below, is complicated by the various gene duplications of PRDM7. Much less variability occurs between the universally conserved patches in the other five genes with a comparable domain, namely PRDM11, PRDM4, PRDM10, PRDM15 and PRDM6. These genes did not experience duplications during placental evolution. The fact that the entire domain is strongly conserved -- with vary different amino acids in each protein -- implies strong selective pressure acts along the entire domain in these five proteins so the 3D structure is not floppy (indeterminate random coil) between the universally conserved patches, and that whatever functions these genes have remained constant during placental evolution.
Note knuckle region in PRDM7/9 has moderate variability. Assuming on analogy with the terminal array zinc fingers that the residues between the second and third zinc ligands contain the residues that provide recognition specificity, these are QNFFIDS. This region has little phylogenetic variability in PRDM7/9. However overall PRDM11, PRDM4, PRDM10, PRDM15 and PRDM6 have even less variability. These regions could bind dna, single stranded rna or another protein involved in regulation. Those these partners may differ, the type of macromolecule will likely be the same because of underlying homology and implausibility of type change. The phylogenetic alignment of non-pseudogenes in the PRDM7/9 group is quite conservative from calJac (new world monkey Callithrix) to human:
PRDM9_homSap YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWLITKGRNCYEYVDGKDKSWANWMRYVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR PRDM9_panTro .......................................................R.......P.....S.....Q.........S.............................E............S......S.......................................... PRDM9_gorGor ...................I.....................................................K........................................................................................................ PRDM9_ponAbe ................................................................................K......................................W.......................................................P.. PRDM9_nomLeu .....................I...T.G...........................................................................................W.......................................................P.. PRDM9_macMul .....................I.....E..................................................Q................................................................................................... PRDM9_papHam ...........................N....................................................K................................................................................................. PRDM7_homSap .....................................................................................S......................S..................................................................... PRDM7_ponAbe .....................................T........................................K................................................................................................... PRDM7_calJac ...I.............................HA.........................................V.......SS............................................................................................ PRDM7_micMur ...K.......................................K.R................E............QV........S....................D..............E...................Q.............................E..TIRQ PRDM7_otoGar ...K.......N..V.........T..E.......V....S..G.RT.......F.........Q..........QV........S....................E.QG...........E....................................................T..Q PRDM7_tupBel ...K.........S.....I..........SL...V.........A.....E.......A.T.............Q.........S....................E.C..............................................E................S.WQ.E PRDM9_oryCun ...K.....L....V....I...............V...............E.......................Q...E.....S....................R.............N............K.......Q..K..........................E..T... PRDM7_oryCun ...K.....L....V....I...............V...............E.......................Q...E.....S....................R.............N............K.......Q..K..........................E..T... PRDM7_ochPri ..........E...V..S..............H..V....S..........E........TT.............QV..E...T.S...........R........P.Q...........N.....................AV.Q.........................E..T... PRDM7_ratNor ...K.........PN....V.....V..R....H.V...............E.............V.......K.Q.........S..................Q.E.Q.........................K............R.....................M..GFT... PRDM7_musMus ...K.........PN....L.....M..R....H.V.........S.....E.............V.........Q.........S..................Q.E.Q.........................K..................................M..GFT... PRDM7_musMol ...K.........PN....L.....M..R....H.V.........S.....E.............V.........Q.........S..................Q.E.Q.........................K..................................M..GFT... PRDM7_dipOrd ...Q......N..TV....I..R.NV.....YD..V.........RQ.S..E........E..............Q.....D...S....M........V........Q...........Y.......................KA.........................R..T... PRDM7_speTri ..DK.....M...PV......I...V.N.D.S.H.T....L.......S..E.........T...........R.Q.........S....................E.Q.................................................................S... PRDM9_bosTau ..QE.........D.............E...A...V.T.....S.KL....E..........H............Q..D.K..I.S...........S........T.L...........HY...........G.......Q.V...............EK....CE.RG.SMFA... PRDM9_oviAri ..QE......N..D.............E...A.....T.....S.RL....E..........H............Q..D.K..V.S....................T.L....................L..QG.......Q.V................D....RD.SG.S..A... PRDM9_munMun ...E......N.............C..E...A.....T..H..S.RL....D.......KV...A........K.Q..DN.....S..A.................T..........................G.......Q.V................DF...RN.RG.S..A... PRDM7_turTru ...K.............A.........E.........T.....S.R.....E.......................Q.........S....................T..............E.....................V..............S.....P...G..SQ.V... PRDM7_lamPac ...K.................................T.......R.....E.........H.............QV........S..........K..........Y...............................................E................S.WQ.E PRDM7_susScr ...K.................................T.......R.....E.........H.............QV........S.........................................................V..............................T..I PRDM7_felCat ...K..........V.........N..G.........T.......R..S..E...............T.......Q.........S....................N................................................................S..ST.K PRDM7_ailMel ...K..........V...............Q......T.......R.............................Q.........S....................N..............E.................................................S..A..K PRDM7_pteVam ...K.............S.I.....E..IR.......T.............E................L......QV........S.....QG.............E.R..............................................................R..T... PRDM7_myoLuc ...K..........V................A.....T.............E........EC...V...Y.....Q.....AI..S....................T.Q..................................V.K.........E...............T.PV... PRDM7_equCab ...N...............I.................T..L....R.....E.......................Q.........S....................I....................................V...........................R..T... PRDM7_sorAra ...N......NK.S...S.I....N..A...S.....T..H..........E....I..................Q..N......S..................V.E.L............Y....................I.K..........................S..T.DK PRDM9_loxAfr ...K.......T..V..A.M.....P..R....H...T..........S..K..........E............QV...K....S.........K..........E..............E....................T.Q.D.....................R.....TS.. PRDM7_choHof ...K.....FEN........LL.....GQ.R.KH...V......L......E.......................QV......T.S......................C.................................A...............................T.EK PRDM9_homSap YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWLITKGRNCYEYVDGKDKSWANWMRYVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR
Central PR(SET) domain descended from PRDM11
Various additional sequences are relevant to understanding the curated placental mammal PRDM7/9 set. For example, the neanderthal genome despite being very far from satisfactory coverage can provide a PRDM9 sequence derived from the human reference sequence using non-synonymous SNPs reported in the corresponding UCSC browser track. The changes reported in the zinc finger domain (R HDL S R) may be enough to have created somewhat of a species barrier, though this involves comparing a fossil sequence to a contemporary human (which are today themselves quite variable). Similarly, the bushman genome sequence might yield an intermediate outgroup, though that assembly (like so many others) remains elusive.
Terminal sequences for 9 additional species of murid rodents have been determined but these have limited value for comparative genomics because they do not even cover the entire terminal exon and their syntenic contexts (and thus homological relationships) were not established. The single individual sequenced may not be representative of the overall population in the zinc finger region (based on the extensive diversity observed in human), diminishing their utility for predicting species barriers. These genes are most likely PRDM7 orthologs only secondarily related to the catarrhine primate PRDM9 set, ie descended from the unique locus present in stem euarchontoglires whereas the latter duplicated from a stem old world monkey PRDM7. It is worth noting that the reported sequences are very orderly and lack the overall chaos of frameshifts and stop codons so often seen in this gene family. The protein accessions are here.
A zebrafish protein put forward as an ortholog to placental mammal PRDM9 seems implausible given that birds, lizard and frog lack notable homologs. It lacks close counterparts in other species of fish with determined genomes and is not syntenic to mammalian gene locations. Thus it might represent an independent gene shuffle that resulted in a similar concatenation of domains (parallel evolution).
The protein lacks the KRAB and SSXRD domains but contains a standard knuckle, PR(SET), early ZNF finger and ZNF repeat domain (all in exons phased identically to human). Although back-blast restricted exons 3-5 to the human proteome has best matches to PRDM9, PRDM7 and the closely related PRDM11 (suggesting orthology of this region), the pre-zinc finger part of exon 6 does not give a clear signal despite its early C2H2 domain, perhaps because of few conserved residues after it. The same could be said for the pre-zinc finger of PRDM9 -- it is apparently just a fast evolving linker region not under selection for amino acid sequence.
While blastp of the zinc finger array is always a problematic exercise, here it gives closer resemblance to other zinc finger genes, notably ZNF658 and ZNF 585, than to any member of the PRDM family. The phase 12 intron here is moderately diagnostic in conjunction with the early zinc finger. The zebrafish terminal zinc finger array, while disorderly, does have several zinc fingers ending in the GEKP-like lockdown cap which supports a relationship with similar caps in PRDM7/9. Genes related to the zebrafish feature are found in salmon, trout, catfish and minnow but not stickleback, fugu, tetraodon or medaka. Transcripts are exceedingly common in contrast to mammals. The missing KRAB and SSXRD domains are believed critical in recruiting other essential proteins to the hotspot in the only systems with experimental data (mouse and human) so this gene cannot fulfill the same functional role.
Thus only central PR(SET) exons are established as directly relevant to the history of PRDM7/9, ie that it was present in the common ancestor of mammals and fish. The terminal exon could represent orthology with extreme relative divergence but the evidence favors a chimeric origin with a different zinc finger terminal exon. The zebrafish gene is thus only a partially verifiable member of the PRDM family, one lacking a convincingly orthologous terminal exon as well as the final fusion with SSX1 as in PRDM7/9. Early diverging tetrapods need re-examination to see if they too have a gene with central PR(SET) exons of PRDM7/9 in a gene with a following phase 12 exon.
Absence in frog, lizard and bird genomes would require persistence through the common ancestor but multiple independent loss events in the descendent lineages. Here frog still has a knuckle-PR(SET) domain most like that of PRDM7/9 but it is attached to a long BED domain and most resembles the human protein ZBED1 family overall.
Even more attractive is the hypothesis that nearest neighbor PRDM11 -- which is highly conserved and effortlessly located in all tetrapods including frog, birds, lizard and platypus -- gave rise to PRDM7/9 via gene duplication. The duplicated gene subsequently neofunctionalized by reciprocal translocation with SSXRD and a ZNF gene to acquire its current N- and C-termini. PRDM11 has hardly changed since the parenting event. Best-blast (of ancestral, consensus, or any individual species) to human proteins is far and away PRDM7/9. This scenario explains -- without multiple gene loss events -- why PRDM7/9 cannot be located in early diverging tetrapods.
Reported PRDM9 orthologs in early diverging bilatera such as Lottia, Capitella and Nematostella can be dismissed as independent occurrences of common ancient domain combinations. None of these domains are mammalian innovations -- PR(SET) traces back to bacterial methylases and zinc fingers also have a long and complex history. Without conservation of all mammalian domains, exon phasing, syntenic chromosomal location and demonstration of descent from a single gene in the last common ancestor, there is no basis for calling such genes orthologous nor assuming they function similarly in meiosis or illuminate mammalian PRDM7/9 evolution. Widespread expression in testes is not supportive as it conflicts with the very restrictive mammalian expression pattern. How could such a fundamental capacity be lost (and replaced by a non-homologous system) so many times in so many other lineages -- all of which have obligatory meiosis?
>PRDM7_danRer Danio rerio (zebrafish) Q6P2A1 transcript BC064665 no KRAB SSXRD or exon 5 but knuckle SET early ZNf C2H2 array 0 MSLSP 1 2 DLPPSEEQNLEIQGSATNCYSVVIIEEQDDTFNDQPF 1 2 YCEMCQQHFIDQCETHGPPSFTCDSPAALGTPQRALLTLPQGLVIGRSSISHAGLGVFNQGQTVPLGMHFGPFDGEEISEEKALDSANSWV 0 0 ICRGNNQYSYIDAEKDTHSNWMK 2 1 FVVCSRSETEQNLVAFQQNGRILFRCCRPISPGQEFRVWYAEEYAQGLGAIWDKIWDNKCISQ 1 2 GSTEEQATQNCPCPFCHYSFPTLVYLHAHVKRTHPNEYAQFTQTHPLESEAHTPITEVEQCLVASDEALSTQTQPVTESPQEQISTQNGQPIHQTENSDEPDASDIYTAAGEISDEI HACVDCGRSFLRSCHLKRHQRTIHSKEKP YCCSQCKKCFSQATGLKRHQHTHQEQEKNIESPDRPSDI YPCTKCTLSFVAKINLHQHLKRHHHGEYLRLVESGSLTAETEEDHT EVCFDKQDPNYEPPSRGRKSTKNSLKGRGCPKKVAVGRPRGRPPKNKNLEVEVQKIS PICTNCEQSFSDLETLKTHQCPRRDDEGDNVEHPQEASQ YICGECIRAFSNLDLLKAHECIQQGEGS YCCPHCDLYFNRMCNLRRHERTIHSKEKP YCCTVCLKSFTQSSGLKRHQQSHLRRKSHRQSSALFTAAI FPCAYCPFSFTDERYLYKHIRRHHPEMSLKYLSFQEGGVLSVEKP HSCSQCCKSFSTIKGFKNHSCFKQGEKV YLCPDCGKAFSWFNSLKQHQRIHTGEKP YTCSQCGKSFVHSGQLNVHLRTHTGEKP FLCSQCGESFRQSGDLRRHEQKHSGVRP CQCPDCGKSFSRPQSLKAHQQLHVGTKL FPCTQCGKSFTRRYHLTRHHQKMHS* 0 >ZBED1_xenTro 0 MQAAEEACAQLEDELL 1 2 FCEDCRLYFRDSCPTHGAPTFILDTPVPENVPSRALLSLPEGLVVKERPQGGFGVWCTIPVIPRGCIFGPYEGDVIMDRSDCTVYSWA 0 0 VRENGSYFYIDASDDSKSSWMR 2 1 YVACASTEEEHNLTVFQYRGKIYYRASQVIPTGTELLVWIGEEYARTLGLKL 1 2 GEHFKYEFGEKELLMKLFQDLQLKPVDSISNHVSSQSQYMCNDMVTPVMQAHRTSYPLNNIGHTSSVFPLLEGTQNLVSLGRAQSRYWTFFGFQGDAYGRIIDKTKIICKLCGVRLSYSGNTTNLRQHLIYKHRRQYNDL
>PRDM11_conSeq consensus of 30 tetrapod PRDM11 orthologs 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKEASGENDVRCINEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNRYKSIDGSDETKANWMR 2 1 YVVISREEREQNLLAFQHSERIYFRACRDIRPGERLRVWYSEDYMKRLHSMSQETIHRNLAR 1 PRDM9_homSap YL Y..M..NF.I.S.AA....T..K.SA.DK.H.N.S..SL.P.LRIGPSGIPQAGLGVW..AL.L.LH......R.TEDEEA.NNY... .TKGR.C.EYV..K.KSW..... ..NCA.DDE....V...YHRQ.FY.T..V....CE.L...GDE.GQE.GIKWGSKWKKE.MA PRDM11_homSap FW FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTSGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWL IVDKNNRYKSIDGSDETKANWMR YVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLAR PRDM11_panTro .. ........................................................................................ ....................... .............................................................. PRDM11_rheMac .. .........................................................I.............................. ....................... .............................................................. PRDM11_calJac .. ........................................................................................ ....................... ....................C......................................... PRDM11_otoGar .. ............................M..................EA...N....I.............................. ....................... ..A...............................R........................... PRDM11_musMus .. ................................................AG.......I.............................. ....................... ..................................R........................... PRDM11_ratNor .. ..............T................................EVG.......I...V.......................... ....................... ..................................R........................... PRDM11_cavPor .. .............................................I.EAG.......I.............................. ...............D......- ..................................R........................... PRDM11_speTri .. ...............................................EA........IS............................. ....................... ..........R.......G...............R......Q...R................ PRDM11_oryCun .. ..........................................L...QEA........I.D.....R.........AA........... .....S................. ........Q.........N.H..........A..R......G.................... PRDM11_ochPri .. ............................M..A...............EA........LSD............................ ....................... ....................H.........Q...R........................... PRDM11_bosTau .. ...............................................EA...N....I.......R...................... ....................... ........S.........................R........................... PRDM11_equCab .. ...............................................EA...N....I.......................T...... ....................... ..................................R........................... PRDM11_canFam .. ...............................................EA...N....I.............................. ....................... ..................................R......................H.... PRDM11_myoLuc .. ..............K....M..........L.................AN..N....I.............................. ....................... ....................H.......................................T. PRDM11_pteVam .. ................................................A...N...SI.............................. ................S...... ....................H.............R........................... PRDM11_eriEur .. ..............K...........................V....EA........I.............................. ......H.........S...... ....................H.............R........................... PRDM11_loxAfr .. ...............................................EA...N....IS............................. ..........V............ .........................V........R......Q.................... PRDM11_echTel .. ............................M..................EG...N....IS....T-..LR......Y.RN......... ....................... ....C...............H.............R.....GQ..................T. PRDM11_dasNov .. ...............................................EA...N....I.............................. ....................... .........................S........Q...............V........... PRDM11_macEug .. ............................M...........P......EA..QN....M.............................. .............T...Q..... ..I..........M......K....V........R.....................Q...T. PRDM11_monDom .. ............................M...........P......EA..Q.....M.............................. ......H......T......... .............M......K....V........R.....................Q...T. PRDM11_ornAna .. ........................................P.I....EA...N....M.............................. .............T......... .I.......................V........R.........................T. PRDM11_galGal .. ........................................P.I....EP...N....M..................S........... .............T......... ..I..........M....................K.....................N...TT PRDM11_taeGut .. ........................................P......EP...N....M..................S..R........ .............T......... ..I..........M................H...K....................MN.SFTS PRDM11_anoCar .. ...................M.L..A...I.........V.P......EAN..R.....G.I....R.Y.....KL.S........... .............T...TS.... ..A..........M...........T........R.....................N...T. PRDM11_xenTro .. ..............S....IL.P..L..I.M.E....SV.C.I.....S...R.....G.I....R.Y.....KL.S........... .............T...TS.... .............M......K....T....Q...K.....................N...TQ
Structural considerations in C2H2 zinc fingers
High resolution structures of C2H2 zinc finger domains have been available for decades. As the name suggests, the divalent zinc atom locks the two cysteines and two histidines into a rigid geometry providing a core conformation that a small peptide of 28 residues could not otherwise stably assume. Note in the unbound state, finger tips must retain flexibility while the domain ensemble scans its genome for specific dna sequences appropriate to its function. Each finger binds a trinucleotide -- in effect making a zinc finger the protein counterpart to tRNA anticodon. However overall binding is not a simple read-off code because adjacent fingers alter each other's specificities in subtle ways.
The linker region TGEKP plays a key role when the correct DNA sequence is encountered, snap-locking its finger down onto its target by capping the C-terminus of its alpha helix. A hydrogen bond between the first threonine and middle glutamate is key to this binding-induced conformational shift. From comparative genomics, it appears that a serine in first position can also form this hydrogen bond. The role of the glycine is to stay out of the way; the lysine counterbalances the negative charge of the glutamate; the proline terminates any helical propensity, allowing a fresh start in the adjacent finger.
While this motif is immensely conserved within C2H2 zinc finger of PDRM9 homologs, exceptions do occur. It is important to understand these because these loss of dna lock-down could loosen or even eliminate trinucleotide binding specificity. Such steps might represent initial stages of pseudogenization. However many exceptions occur within the first or last fingers. It is also common for fragmentary and imperfect motifs to end the protein, sometimes continuing on in another reading frame past the current stop codon.
Note in aligning zinc finger motifs, the breaks should always be put at the end of the linker region. It is completely illogical to break at the first cysteine as some authors do because capping by the linker region is specific to its zinc finger, not the following one.
Predicting dna binding sites of zinc finger domains
Origin of Species and all that
The origin of species is an old and exceedingly complex topic. PRDM7/9 cannot provide a unified molecular basis for it because orthologs do not exist outside of mammals. Yet genetic mapping shows PRDM7 at the core of hybrid sterility in mice (a enticing proxy for speciation) and PRDM9 sites are key to meiotic recombination in humans.
Although placental mammals very greatly in the specifics of PRDM7/9 gene expansion, some variation on the mouse/human might still be applicable. However that cannot be the sole explanation even here because some species such as dog lack any functional gene family member. An Oct 2011 paper established that the PRDM7 gene of the boxer used for the genome project is in fact representative of dog breeds; wolves, jackals and foxes also have the same inactivating frameshifts. Other carnivora such as mink -- while possessing a seeminly intact PRDM7 gene -- lack a sufficiently long zinc finger array to specify hotspots. Some other mechanism is needed to mark the sites of double stranded breaks that initiate meiosis in these Carnivora.
It is problematic whether the PRDM7/9 mechanism extends to marsupials and monotremes and not an option for earlier amniotes such as birds which lack any semblance of the gene (though homologs for its parts occur). Conceivably, a zinc finger array from a different gene family steps in. However drosophila utilizes very different gene products to control meiosis (with other genes such as DMRT1 more universal). There cannot be a unified molecular basis for speciation within bilaterans based on zinc finger proteins.
Thus despite being an ancient core process, the meiotic machinery is unexpectedly not conserved at the gene family level, instead exhibiting discontinuities in some gene families utilized to accomplish it. That is not unlike SRY sex determination (which underwent abrupt changes in marsupials and placentals shortly after divergence from monotremes), or the sex chromosomes themselves (which are homologous to bird but with hetergametic XY males in platypus and taken from different autosomes in therans).
PRDM7/9 do affect an immense number of other important topics in mammalian evolution through their specification of meiotic recombination sites. Because the crossover bottleneck in pseudoautosomal regions, these influences become deeply intertwined with special aspects of sex chromosome evolution. Moving beyond the narrow perspective of PRDM7/9 evolution, the following issues can be explored:
- meiotic sex chromosome inactivation (MSCI)
- progressive chromosome Y degeneration or conversely maintainence
- gene dosage compensation for chromosome X
- sex determination via SOX3 evolving into SRY on chromosome Y after platypus divergence
- co-evolution with key meiotic regulatory genes such as DMRT1
- expansion and contraction of the pseudoautosomal region in placentals
- barriers to introgression during hominid history
- impacts on lineage sorting within great apes
- boundaries and persistence of haploblocks
- localization of gene conversion
- random X-inactivation in females
- XY bodies and chromatin structure during meiosis
- evolutionary strata in sex chromosomes due to inversions reducing recombining region
- translocations and other chromosomal rearrangements in evolution and disease
These topics will be further developed in October.
(to be continued)
Supplemental information
The sections below store data used above. This includes curated sequences from all available mammals for PRDM7 and PRDM9 and additional their partial paralogs in the PRDM gene family. These latter have extensive comparative genomics alignments readily available elsewhere (UCSC genome browser, under GeneSorter feature and ProteinFasta feature in gene details page) so that is not repeated here.
While this topic has a long history in the peer-reviewed scientific literature, only the most recent articles are provided here because their reference sections satisfactorily summarize pre-2005 studies. Instead, the focus here is identifying free full text access to the recent articles, preferably as html which better supports copying snippets of text. The journal, google, and PubMed all provide forward citations to still other articles that cite the articles provided here.
Curated reference sequences
The sequences below have largely been compiled from genome projects -- only rarely do validating transcripts exist at GenBank. Sequences with a single frameshift or other glitch have been edited in some cases to show full length proteins on the theory that the error either reflects an atypical individual chosen for sequencing, sequencing error in low coverage projects within a difficult region, one allele in balanced polymorphism, or a mutant allele. However such sequences may instead reflect early stages of pseudogenization. Other sequences are in fact clearly pseudogenes; here recognizable exons are represented to allow determination of historic repeat number and rough dating of loss of function.
Chimp PRDM9 has a thoroughly garbled 11th repeat (3 frameshifts, nnnnn in the CGSC 2.1.3/panTro3 Oct 2010 assembly) followed by eight additional repeats. It is difficult to say if these are still in the initial reading frame. No data is currently available for Pan paniscus but despite minimal coverage, Pan troglodytes schweinfurthii has a long trace read in this region (ti|2009092447) that covers repeats 3-11 sharing the first frameshift in Pan troglodytes but soon degenerating into unmistakable pseudogene (ie the frameshifts in Pan troglodytes are not sequencing artifacts):
PRDM9 Pan troglodytes schweinfurthii
THTGEKP 3
YVCRECGRGFSVKSSLLSHQSTHTGEKP 4
YVCRECGRGFSVKSSLLSHQRTHTGEKP 5
YVCRECGRGFSVKSSLLSHQRTHTGEKP 6
YVCRECGRGFSVKSSLLSHQRTHTGEKP 7
YVCRECGRGFSQQSHLLSHQRTHTGEKP 8
YVCRECGRGFSQQSHLLSHQRTHTGEKP 9
YVCRECGRGFSQQSHLLSHQRTHTGEKL 10
YVCRECGRGFSRAVTPPQTPETHTGEKL 11
YVCREcGRGFSDKsSLlSVTRVHTQGRA 12
YVCRECGRGFSWQS 13
Gorilla, despite its importance, has a poor quality second assembly that is riddled with gaps and misplaced contigs. The PRDM9 gene terminates early in the zinc finger array with a small gap before a misplaced LINE element. The neighboring genes are not plausible though PRDM9 is correctly placed on chr5. PRDM7 in gorilla has four frameshifts in the last exon. Those are 'restored' below to reveal gene history (which in this case is 3.5 repeats with no trailing debris). Lower case letters indicate ends of correct reading frames.
Sumatran orangutan PRDM9 has a distal zinc finger array frameshift in contig ABGA01214983 (aGGGGAGAAG CCCTATGTCT GCAGGGAGTG) which also appears in the July 2007 assembly provided by UCSC (WUGSC 2.0.2/ponAbe2). However there is no support whatsoever for this extra adenosine in the underlying raw Sanger trace reads used to make the contig -- all reads omit it and have the expected reading frame. Consequently, the assembly appears to be in error. The full length sequence is provided here. Orang PRDM9 has the expected syntenic location between CDH10 and CDH12.
Gibbon has a stop codon in exon 6 that is supported by all 6 of the available Sanger traces.
In the case of more intensively studied species such as human and mouse, the number of C2H2 repeats varies widely. Only the reference sequence representative is shown here. This variation likely occurs in all species with the individual animal chosen for sequencing not necessarily the most common allele. Many clades have independent histories of gene amplification and gene loss, making both orthologous and functional comparisons problematic at substantial divergence.
These sequences are under constant revision as anomalies surface and are resolved by re-examining the original Sanger trace reads. However when coverage is low, it is very difficult to distinguish read error from actual oddities. For example, the horse genome assembly shows a disabling frameshift in seventh exon. That is apparently based solely on trace ti|1206069852 and arises from a truncated homopolymer run error CCCC that is frame-preserving CCCCC in ti|1330418597, ti|1322386025 and ti|1288100157. These latter reads however later have problems of their own. Another issue is individual traces covering part of a zinc finger repeat -- these cannot always be assigned reliably to PRDM7/9 as many similar zinc finger repeats exist in these genomes.
Other useful sequences such as PRDM11, PRDM4 and zinc finger semi-homologs having similar exon and domain structures, are provide in the subsequent section along with syntenic markers such as GAS8.
>PRDM9_homSap Homo sapiens (human) genome Prim gene 13 CDH12 chr5 10 exon size 18,301 bp KRAB SSXRD SET C2H2 0 MSPEKSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKL YVCRECGRGFSWKSHLLIHQRIHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRDKSHLLRHQRTHTGEKP YVCRECGRGFRDKSNLLSHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRNKSHLLRHQRTHTGEKP YVCRECGRGFSDRSSLCYHQRTHTGEKP YVCREDE >PRDM9_panTro Pan troglodytes (chimp) 2010 assembly exon 8 in gap, 11th repeat 3 frameshifts, only 1 trace supports stop codon *VCREDE ti|451784509 0 MSPERSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPLMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELKKKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYKGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPRSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTAKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKP YVCRECGRGFSWKSHLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHRTTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSQQSNLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSKQSHLLSHQRTHTGEKP YVCRECGRGFSVQSNLLSHQRTHTGEKL YVCRECGRGFSQQSHLLRHQRTHTGEKP YVCReCGRgFSVkSSLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSKQSHLLSHQRTHTGEKP YVCRECGRGFSQQSHLLSHQRTHTGEKP YVCRECGRGFSQQSHLLRHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECERGFSQQSHLLRHQRTHTGEKP YVCRECGRGFSRQSALLIHQRTHTGEKP *VCREDE >PRDM9_gorGor Gorilla gorilla (gorilla) CABD02290262 CABD02290264 cdh12 chr5 ZNF at end of contig Aug 2009 assembly poor quality 0 MSPERSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPCMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRKKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPIFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYKGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARTLLQPENPCPGDQNQEQQYPDPRSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKP YVC >PRDM9_ponAbe Pongo abelii (Sumatran orangutan) CDH10- PRDM9+ CDH12- distal frameshift aGGGG --> GGGG causing loss of 1.5 repeats in ABGA01214983 0 MSPERSQEESPEDDTERTERKPT 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 0 0 GMPKASFNNESSLKELSETANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRSKETEGNTYSLRERKGHAYKEISEPQDDDYL 1 2 CEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALTLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITKDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNHEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKP YVCRECGRGFSRQSVLLIHQRTHTGEKP YVCRECGRGFSRRSVLLIHQRTHTGEKP YVCRECGRGFSQQSVLLIHQRTHTGEKP YVCRECGRGFSRRSVLLIHQRTHTGEKP YVCRECGRGFSWKSVLLRHQRTHTGEKP YVCRECGRGFSQQSVVFIHQRTHTGEKP YVCRECGRGFSGKSVLFRHQRTHTGEKP YVCRECGRGFSDKSGVCYHQRTHTgEKP YVCRECGRGFSVKSNLLSHQRTHTEEKL YVCREDE* >PRDM9_nomLeu Nomascus leucogenys (gibbon) ADFV01015315 Prim gene 10 cdh12 ADFV01015317 ADFV01015319 no synteny CpG stop exon 6 in 6/6 traces VCRKDE* in altered reading frame 0 MSPERSQEESPEEDTERTEQKPT 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRMEQRKHQK 0 0 GMPKASFSNESSLKELSGAANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRRKETEGKMYSL*ERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFTDSCAAHGPPTFIKDSTVGKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCQVIRPGCEPLVWYGDEYGQELGIKWGSKWKKELTAER 1 2 EPKAEIHPCPSCCLAFSSQKFLSQHVARHHSSQNFPGPSARKFLQPENPCPGDQNQEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWQREISRAFSSSPKVQMGSCRVGKRIIEEESRTGQKVNPGNTGQLFVGVGISRIAE VKYAECGQGFSDKSDVITHQRTDTGEKP YLCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSKKSNLLSHQRTHTGEKP YVCRECGRGFSDKSSLLRHQRTHTGEKP YVCRECGRGFSQKSSLLSHQRTHTGEKP YVCRECGRGFSQKSSLLSHQRTHTGEKP YVCRECGRGFSDKSSLLRHQRTHTGEKP YVCRECGRGFSQKSSLLSHQRTHTGEKP YVCRECGRGFSVKSNLLSHQRTHTGEKP YVCRECGRGFSDKSSLLRHQRTHTGEKP *vCRKDE >PRDM9_macMul Macaca mulatta (rhesus) genome Prim gene 9 CDH12 chr6 exon 4 lost to Ns 0 MSPERSQEESPEEDTERTERKPT 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAVRVEQSKHQK 0 0 GMPKASFNNESSLKEVSGMANLLNTSGSEQAQKPVSPPGEARTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFIKDSAVEKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITQDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSTQNFPGPSARRLFQPENLCSGDQNQEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWPKEISRAFSSPPKGQMGSSRVGERMMEEEYRTGQKVNPENTGKLFVGVGISRIAK VKYGECGQGFSDKSDVIIHQRTHTGEKP YLCRECGRGFSQKSSLRRHQRTHTGEKP YLCRECGRGFRDNSSLRYHQRTHTGEKP YLCRECGRGFSNNSGLCYHQRTHTGEKP YLCRECGRGFSDNSSLHRHQRTHTGEKP YLCRECGRGFSNNSGLRYHQRTHTGEKP YLCRECGRGFSNNSGLRHHQRTHTGEKP YLCRECGRGFSQKANLLRHQRTHTGEKP YLCRECGRGFSQKADLLSHQRTHTGEKP *vCRKDE >PRDM9_macFas Macaca fascicularis (crab-eating macaque) CAEC01530962 CAEC01530970 frameshift exon 7 fragmentary array 0 MSPERSQEESPEEDTERTERKPT 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 0 GMPKASFNNESSLKEVSGMANLLNTSGSEQAQKPVSPPGEARTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFIKDSAVEKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFgPYEGRITQDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSHHVERNHSTQNFRGPSARRLLQPENLcSGDQNQEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWQKEISRAFSSPPKGQMGSSRVGERMMKEEYRTGQKVNPENTGKLFVGVGISRIAQ VKYGECGQGFSEKSDVIIHQRTHTGEKP YLCRECGRGFSRKSNL >PRDM9_papHam Papio hamadryas (baboon) genome Prim gene 11 cdh12 contigs scattered 0 0 0 1 2 1 2 VKPPWMAFRVEQSKHQK 0 0 EMPKTSFSNESSLKELSGTPNLLSTSGSEQAQKPASPPGEASTSGQHSRLKL 1 2 ELRRKEAEGKMYSLRERKGHAYKEVSELQDDDYL 1 2 ycEMCQNFFIDSCAAHGPPTFVKDSAVNKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDKEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSTQNFPGPSARRLLQPENLCSGDQNQEQQYSDPCSCNDKTKGQEIKERSKLLNKRTWQKEISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPENIGKLFVEVGISRIAK VKYGECGQGFSDKSDVVIHQRTHTREKP YLCRECGRGFSQKSNLLRHQRTHTGEKP YLCRECGRGFRDNSSLRCHQRTHTGEKP YLCRECGRGFRDNSSLRCHQRTHTGEKP YLCRECGRGFSDNSSLRYHQRTHTGEKP YLCRECGRGFRDNSSLRYHQRTHTGEKP YLCRECGRGFSVKSNLLSHQRTHTGEKP YVCRECGRGFSDNSSLRCHQRTHTGEKP YLCRECGRGFSQMSHLRCHQRTHTGEKP YLCRECGRGFSVKSNLLSHQRTHTGEKP YVCRECGRGFSRKANLLSHQRTHTGEKP *vCRKDE >PRDM7_homSap Homo sapiens (human) pseu chr16 TUBB3+ DEFB+ AFG3L1+ DBNDD1- GAS8+ PRDM7- 4 frameshifts 0 MSPERSQEESPEGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKMNYNALITV 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAFRGEQSKHQK 0 0 GMPKASFNNESSLRELSGTPNLLNTSDSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKGHAYKEISEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSSANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWSGDEYGQELGIkWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQERQYSDPRCCNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVITHQRTHTGGKP YVCRECGRgFSRKSDLLSHQRTHTGEKP YVCRECERGFSRKSVLLIHQRThTGETP vCRKDE >PRDM7_panTro Pan troglodytes (chimp) genome Prim pseu 2 GAS8+ chr16 0 MSPERSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPLMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELKKKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYKGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLQPENP*PGDQNQERQYSDPRCCNDKTKGQEVKERSKLLNKWTWQREISRAFSSLPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVITHQRTHTGGKP YVCRECGQGFSRKSVLLIHQRTHRGEKP *vCRKDE >PRDM7_gorGor Gorilla gorilla (gorilla) genome Prim pseu 3 GAS8+ chr15730 four frameshifts in last exon 0 MSPERSQEESPEGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATQPVFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 0 0 GMPKASFNNESSLKELSGTPNLLNTSGSEQAQKPVSPPGEASTSGQHSRRKL 1 2 ELRRKETEGKMYSLRERKGHAYKEISKPQDDDYL 1 2 yCEMCQNFFIDSCAAHGPPTFVKDSAVDKRHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDKEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVALQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQERQYSDPRCCNDKTKGQEIKERSKLLNKrTWQReISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK YGECGQGFSWKSNLLRHQRTHTGGKP YVCRECGRGFSWKSDLLSHQRTHTGEKP YVCRECGRGFSWKSNLLSHQRTHTgEKP *vCRKDE >PRDM7_ponAbe Pongo abelii (Sumatran orangutan) genome-blat same late stop codon as chimp *VCRKDE* no cryptic repeats, adjacent GAS8 0 MSPERSQEESPKGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWTEMGDWEKTRYRNVKRNYKTLITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAFRGEQSKHQK 0 0 GMPKASFNNESSLKELSGTQNLLNTSGSEQAQKPVSPPGEASTSGQHSTLKI 1 2 ELRRKETEGKTYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDKEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCAWDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMPGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARHLLQAENPCPGDQNQEQQYSDPDCCNDKTKGQEIKERSKLLNKRTWQREISRAFSSSAKGQMGSSRVGERMMEEESGTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVITHQRTHTGGRS YICRESGRGFTQKSGLLSHQRTHTGEKP YVCRECGWGFSQKSNLLRHQRTHTGEKP YVCRECGRGFSRKSVLLIHQRTHTGEKP *vCRKDE >PRDM7_nomLeu Nomascus leucogenys (gibbon) ADFV01125891 pseu gas8+ synteny implied by non-coding 0 0 0 1 2 1 2 IKSPWMAVRVEQSKHQK 0 0 GMPKASFNNESGLKELSGSQnLLNTSG*EQARKPVSPPGEASTSGQHSRQKL 1 2 ELRRKETEGKMYSL*ERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFTDSCAAHGPPTFVKDSAVDKGHPNHSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKS*ANWMK 2 1 YVNCARDHEEQNLVAFQYHRQIFYRTCQVIRPGCEPLVWYGDEYGQELGIKWGSKWKKELTAER 1 2 EPKPEIHPCPSCCLVFTSQKFLSQHVECNHSSQNFPGPSARKLLQRENPCPGDQNQEQQYSDSRSCNDKTKGQEIKERSKLnKRIWQRKISRAFSSLPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVIAHQGTHTGGKS *ICRECGWGFSQESHLLIHQRTHTGEKL YVCRECGQGFSQKSDLLSHQRTHTGEKP YVRRECGRGFSQKSNLLSHQRTHTEEKP YVCRECGWGFSQKSHLLIHQRTHTGKKP *vCRKDE >PRDM7_macMul Macaca mulatta (rhesus) pseu GAS8+ stop codon exon 5 in 6 of 6 trace reads, frameshift in exon 10 0 0 0 1 2 1 2 VKPPWMAFRVEQSKHQK 0 0 EMPKTSFNNESSLKELSGTPNLLSTSDSE*AQKPASPPGEASTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKRHAYKEASELQHDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDNAVNKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPCEGRITEDKEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSWAKWMR 2 1 1 2 EPKPEIYPCPSCCLAFSSQKFLSqHVERNHSSQNFPGPSARKLLQSENPCPGDQNQEQQYSDPSSCNDKTKGQEIKERSKLLNKRTWQREILRAFTSPPKGQMGSSRVGERMMEEEFRTGQKANPGNTGKLFVGVEISRIAK VKYGECGQGFSGKSDVITHQRTHTEGKP YVCRGCGRRFSQKSSLLRHQRTHTGEKP *vCKKNE >PRDM7_macFas Macaca fascicularis (crab-eating macaque) CAEC01352986 CAEC01352983 4 frameshifts 1 stop codon 0 0 0 1 2 1 2 0 0 EMPKTSFNNESSLKELSGTPNLLSTSDSE*AQKPASPPGEASTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKRHAYKEVSELQHDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDNAVNKGHPNRSALSLPPGGlRIGPSGIPQAGLGVWNEASDLPLGLHFGPCEGRITEDKEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 1 2 EPKPEIYPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQSENPCPGDQNQEQQYSDPSSCNDKTKGQEIKErSKLLNKRTWQREILRAFTSPPKGQMGSSRVGERMMEEeFRTGQKaNPGNTGKLFVGVEISRIAK VKYGECGQGFSGKSDVITHQRTHTEGKP YVCRGCGRRFSQKSSLLRHQRTHTGeKP *vCKKNE >PRDM7_papHam Papio hamadryas (baboon) genome Prim pseu 2 gas8+ contigs scattered 0 MSPERSQEESPEEDTERTEWKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAVRVEQSKHQK 0 0 GMPKASFNNESSLKEVSGMANLLNTSGSEQAQKPVSPPGEARTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFIKDSAVEKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITQDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIYPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQSENPCPGDQNQEQQYSDPSSCNDKTKGQEIKERSKLLNKRTRQRQILRAFTSPPKGQMGSSRVGERMMKEEFRTGQKANPGNTGKLFVGVEISRIAK VKYGECGQGFSDKSDVVIHQRTHTREKP YVYRgCGQGFSIKSNLLRHQRIHTGEKP >PRDM7_calJac Callithrix jacchus (marmoset) genome Prim gene 12 GAS8+ chr20 one frameshift in repeat area chr20 terminus 0 MSPERSQEESPEGDTGRTEQKPM 0 0 VKDAFKDISMYFSKEEWAEMGDWEKTRYRNMKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPGMAFRVGQSKHQK 0 0 GMPKASFGNESSLKKLSGTANVLNTSGPEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRRKDTEEKMYSLRERKGLAYKEVSEPQDDDYL 1 2 yCEICQNFFIDSCAAHGPPTFVKDSAVDKGHPNHAALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRVTEDEEAASSGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 ESKPEIHPCPSCCLAFSSQKFLSHHVERNHSSQNFPGTSTRKLLQPENPCPGKQKEEQQYFDPCNSNDKTKGQETKERSKLLNIRTWQREMARAFSNPPKGQMGSSRVEERMMEEESRTGQKVNPVDTGKLFVGVGISRIAK AKYGECGQGFSDMSDVTGHQRTHTGEKP YVCRECGRGFSQKSALLSHQRTHTGEKP YVCRECGRGFSQKSHLLSHQRTHTGEKp YVCTECGRGFSQKSVLLSHQRTHTGEKP YVCTECGRGFSRKSNLLSHQRTHTGEKP YVCRECGRGFSRKSALLSHQRTHTGEKP YVCRKCGRGFSQKSNLLSHQGTHTGEKP YVCTECGRGFSQKSHLLSHQRTHTGEKP YVCRKCGRGFSQKSNLLSHQRTHTGEKP YVCRECGRGFSFKSALLRHQRTHTGEKP YVCRECGRGFSRKSHLLSHQGTHIGEKP YVCRECGRGFSRKSNLLSHQRIHTGEKP YVRREDE >PRDM7_saiBol Saimiri boliviensis (squirrel_monkey) AGCE01149118/AGCE01147692/AGCE01012341/AGCE01145199 fragments no synteny but not in cadherin 10/12 complex MSPERSQEESP GDTGRTEQKPM VKDAFKDVAIYFSKEEWAEMGDWEKTRCRNVQRNYNALITI GLRATQPAFMCHRRQASKLQVDDTEDSDEEWTPRRQ RPVSPPGEASTSGQHSRVKP YVNCARDDEEQNLVAFQYHRQIFYRTCQVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR EPKPEIHPCPSCCLAFSSQKFLSRHVERNHSSQNFPGTSTRKLLQPENPCPGKQKEEQRYFDPCNSNDKTKGQEIKERSKLLNTRTWQREIARAFSYPPKGQMGSSRVEERMMEGESRTGQKVKPVDTGKLFVGVGISRIAK ANYGECGQGFSGMSDVTAQQRIHTGEKP YVCRECGRGFGHKSTLLSHQRTHTGEKP Y >PRDM7_tarSyr Tarsius syrichta (tarsier) ti|1493610848 ABRT011082008 ti|1633244849 ABRT010499286 ABRT010929713 ti|1646504284 pseu gas8? double frameshift in exon 5 single trace ti|1623660607 0 MSPDRSPEDSPEGDTGRTECKSA 0 0 VKDAFKDISIYFTKEEWAEMGDWEKIRYRNVKRNYNTLIAI 1 2 GLRAPRPAFMCHRKRAIKPLVDDTEDSDEEWTPRQQ 1 2 VKPPWMAFRVEQNKHQK 0 0 GMPRAPLSIVSSLKELSEMANLLnTSDSEQAWKPVSPSrEASTSEQHSRKKI 1 2 EFRKKEIEVNMYSLRERKDCAYKEVNEPQDDDYL 1 2 YCEQCQNFFIDSCATHGIPTFINDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASELPLGLHFGPYEGQITDDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRIIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 ETKPKILPCSSCSLALSSQKFFSQHVKCNHPPQIFPGTSARKYVQPENPCPEDLNQEQQQSDPHSWNDKSKCQEAKERSKPMHKRTQQRESSRSLSNPPTEQTESSREKERMMKEEPSTSQKVHLGDTGYEFHHIGASAAR >PRDM7_micMur Microcebus murinus (lemur) ABDC01433247 ABDC01371462 gas8+ weak coverage last exon has two frameshifts 0 MSPNKSQEESPEVDAGRTGWKPT 0 0 DKDAFKDISIYFSKEEWAQMGDWEKIRYRNVKRNYNVLITI 1 2 GLRAPRPAFMCHRRQAIKHQVDDTEDSDEEWTPRQQ 1 2 0 0 1 2 SEPQDDDYL 1 2 YCEKCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLKIRPSGIPQAGLGVWNEASELPLGLHFGPYEGQVTEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDDSWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCQVIRPGCELLVWYGDEYGQELGIKWGSKWKEELTIRQ 1 2 EPKPEIHPCPSCSLAFSSQKFLSQHVKHTHSSQISPRTSGRKHLQPENPCPGDQNQEQQHSDPHSCNDKAKDQEVKERPKPFHKKTQQRGISRAFSSPPKGKMGSCREGKRIMEEEPRTGQKVGPGDTDKLCAAGGISRISR VKYGDSGQSFSDKSNVIIHQRTHTGEKP YVCRECGRGFSQKSDLLKHQRTHTGEKP YVCRECGRGFSQKSHLLRHQRTHTGEKP YVCRECGRGFSQKSDLLIHQRTHTGEKP YVCRECGRGFSCKSHLLIHQRTHTGEKP YVCRECGRGFSCKSSLLIHQRTHTGEKP YVCReCGrGFSRKSDLLIHQRTHTGEKP CVCRKGE >PRDM7a_otoGar Otolemur garnettii (galago) AAQR03189271 (43912 bp) adjacent ARFGEF1 not GAS8 0 MSPNRSQEESPE 0 0 VRGAFKDTYKYFSMEELAEMGDWEKIHHGNVERNYDVLIDR 1 2 GLRAPQPAFMGHRRQAIKYQVDDTEDSDEEWTPRQQ 1 2 GKHSLMAFRMQPRKRQK 0 0 GMPRAPLSHDSILQDLSGPANSLNISDSEQHQNCVSLPGEANASGQNSRRKS 1 2 ALRRKEIEARSYNLRERTDRSYEEVSEPQNDDYL 1 2 YCEMCQDYFIDRCDVHGPPTFVKDIAVDKGHPNRAALTLPPGLSIRQSGIPQAGDGVWNEACELPLGLHFGPYEGQVLEDEEAARSGYAWK 0 0 ITKGRNCYEYVDGKDQSQGNWMR 2 1 YVNCARDDEEQNLVAFQYHSQIFYRTCRVIRPGCELLVWYGTEYGKKLGIMWTSKRKKELTGQ 1 2 DPKPENHPCPSCSLAFSSQKSLSQHVEGTHSSQIFPGTSVRKHCRPEHLYPGDQNQEQQLSDPHDQNDKTKGQEMNEISKTSQEKTQQSSISGISSHTPEGQMGNSRDSERMVEPGQNMGPGETGKLCVKVEISRIVK VANGQCGQEFSQTSNLHTHQRTHTGEKP YVCSQCGHGFRYKSNLLTHQRIHTGEKP YICTECGQQFRQTSNLLAHQRIHTGEKP YVCSDCGKGFSQKSNLRTHQKTHTGEKA YVCSECGKGFTRKENLLIHHRTHTGEKP YICSDCGKRFSQKSNFLTHQKTHTGEKA CVCRECGKGFSRKATLLIHQRTHKGEkP YVYRSCGQIFIHKSNLNRREKTHTGEKS >PRDM7b_otoGar Otolemur garnettii (galago) AAQR03144890 (36951 bp) adjacent GAS8+ pseudogene 0 0 0 1 2 GLRAPRPAFMCHRRQATKYKVDDTEDSDEEWTPRQQ 1 2 VKHPWMAFRMEQSKRQK 0 0 ILKKCMLSFNMHLKELSGPASLPNISGSEQHQKHMSSPREASTSGQHSGRKS 1 2 DLRIKEIEVRMYSLRERKGHAYKEVSEPQDDDYL 1 2 yCEKCQNFFIDNCAVHGPPTFVKDTAVEKGHPNRSVLSLPSGLGIRTSGIPQAGFGVWNEASDLQLGLHFGPYEGQVTEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDESQGNWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGQ 1 2 EPKPEIHPCPSCSLAFSTQKFLSQHVERTHPSQISQGTSGRKNLRPQTPCPRDENQEQQHSDPNSRNDKTKGQEVKEMSKTSHKKTQQSRISRIFSCPPKGQMGSSREGERMIEEEPRPDQKVGPGDTEKFCVAIGISGIV KVKNRECVQSFSNKSNLRHQRTHTGEKP YMCRDCGRGFSHKSSLFRHQRTHTGEKP YVCRDCGRGFSLKANLLTHQRTHTGEKP YVCRDCGQGFSQKAHLLRHQRTHTGEKP YMCRDCGQGFSRKAYLLTHQRTHTGEKP YVCRDCGQGFSQKAHLLTHQRTHTGEKP YVCRDCGRGFSHKSSLFRHQRTHTGEKP YICRDCG*SFRDRSNLLRHQRTHTGEKL YVCRECGQGFNLKVTLLTHQRTHTGEKP YVCRDLG*SFHNRSNLLTHqRTHIGEKP YVCRDFGRGFSQKAHLLTHQR >PRDM7_tupBel Tupaia belangeri (tree_shrew) AAPY01316756 ti|1061183379 ti|1061388949 ti|1076935836 AAPY01523531 ti|1074533669 AAPY01523530 no synteny QVTSDKTYFSGQMEDKSGHHTPP* runout 0 MRRYKSPEESPEGDAGRTEWKPT 0 0 VKDAFKDISVYFSKEEWAQMGEWEKIRYRNVKRNYTTLIAI 1 2 GLRAPRPAFMCHRKLAVKPHMDDAEDSDEEWTPRQQ 1 2 0 0 1 2 GKMYSLRERKCGTYKEVHEPQDDDYL 1 2 yCEKCQNFFIDSCSAHGPPIFVKDSAVDKGSLNRSVLSLPPGLRIAPSGIPEAGLGVWNAATDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDESCANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGEEYGQELGIKWGSKWKKSLWQGE 1 2 EPRPEIHPCLSCSLAFSSQKFLNQHVEHNHSCQRVPETSARKHLHPEHTHPGDEIQEQHHSDPQHCSDKADYEKVKERSKPSQERTRQRTMSRTFCSPCKGQMENAREGERMTGQRTDQKVGAEDTGKLLVRVGSSRIAG VKYGECGQGLINKSNASRHQRTHTGEKP YGCRECGRGFSQQSDLIRHQRTHTGEKP YLCGECGRGFSLQSSLIRHQRTHTGEKP YLCGECGRGFSRQSHLIIHQRTHTGEKP YVCRECGRGFSLQSNLIIHQRTHTGEKP YGCRECGRGFSQQSSLIRHQRTHTGEKP YLCGECGRGFSRQSHLIIHQRTHTGEKP YVCRECGRGFSLQSNLIIHQRTHTGEKP YGCRECGRGFSQQSSLIRHQRTHTGEKP YVCRECGRGFSRHSSLIIHQRTHTGEKP YLCGECGRGFSRQSHLIIHQRTHTGEKP YVCRECGRGFSQQPQLIIHQRTHTGEKP YVCRECGRGFRCQSHLIIHQRTHTGEKP YVCRECGRGFSQQPHLIIHQRTHTGEKP *VCRKGE >PRDM7_oryCun Oryctolagus cuniculus (rabbit) genome Glir gene 8 other Un0161 exon 2 ttt to tt restores frame; ZNF717+ DCAF4+ YAP1+ PRDM9- qTer 0 MSAAAPAEPSPGADAGQARGKPE 0 0 VQDAFRDISIYFSKEEWAEMGEWEKIRYRNVKRNYCALVAI 1 2 GLRAPRPAFMCHRRLAVRARADDTEDSDEEWTPRQQ 1 2 VKPPWMAFRTEHSKHQK 0 0 GMPRLPVNNESSLKELSGTANLLKTTGSEEDQKPSFPPKETRTSGQHSTRKL 1 2 GLRRKNIEVKMYSFRKRKSQAYKECSEPQDDDYL 1 2 YCEKCQNFFLDSCAVHGPPIFVKDSAVDKGHPNRSVLSLPPGLRIGPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEEEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDRSWANWMR 2 1 YVNCARNDEEQNLVAFQYHKQIFYRTCQVIKPGCELLVWYGDEYGQELGIKWGSKWKEELTAGR 1 2 EPKPEIHPCPSCSLAFSSHKFLSQHMERSHSSQIFPGAPARNHLQPANPCPGKEHQKLSDPQSWNDKNEGQDVKEKSRFSSKRTRQKAISRSFSSLPKGQVETSREGERMIEEEPRIGQELNPEDTGKSSVGAGLSRIAG VKYRDCRQGLSDKSHLINGQRAHTGEKP YACRECERGFTVKSNLISHQRTHTGEKP YACRECGRGFTVKSALTTHQRTHTGEKP YACRECGRGFTVKSHLISHQRTHTGEKP YACRECGRGFTVKSALITHQRTHTGEKP YACRECGQGFTVKSNLISHQRTHTGEKP YACRECGRGFTQKSHLINHLRAHTGEKP YACRECGRGFTVKSDLISHQRTHTGEKP YACRVDE >PRDM7_ochPri Ochotona princeps (pika) ti|1534455888 AAYZ01312269 AAYZ01242582 no synteny 0 0 0 VPDAFRDICVYFSRAEWAEMSESEKLRYRDVKRNYSALRAI 1 2 GFRAPRPAFMCRRRLAGRAREEDTDDSDEECTLRPQ 1 2 VKPPWMAFRTEHSKHQK 0 0 GIPKVPTHHESSVTEAPRTAPFLRPAGSQQGRKPAFPPEEASASGQHSTRRL 1 2 EGRRRKTDLKIYSLRKRKSRTYKECSEPQDDDYL 1 2 YCEMCQNFFIESCAVHGSPTFVKDNPVGKGHPHRSVLSLPSGLRIGPSGIPEAGLGVWNETTDLPLGLHFGPYEGQVTEEEEATNSGYSWL 0 0 ITKGRNRYEYVDGKDPSQANWMR 2 1 YVNCARNDEEQNLVAFQYHRQIFYRTCRAVRQGCELLVWYGDEYGQELGIKWGSKWKEELTAGR 1 2 EPKPEIHPCPSCSLAFSSHKFLSRHTERSHSSQVFPKTSPRRLQPVNPCRGTEHREPLDPHSWNDTAEGQEVIENSSSVSTRTRQRQISAAFCSLRKGQVEASREGERKAEESPRINQEVNAQDTAKSSVRAGLPSRVT VKCGDCRQDL >PRDM7_ratNor Rattus norvegicus (rat) P0C6Y7 Glir gene 10 PDCD2 chr1 FM103467 single transcript from body fat 0 MNTNKPEENSTEGDAGKLEWKPK 0 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 2 GLRAPRPAFMCYQRQAIKPQINDNEDSDEEWTPKQQ 1 2 VSSPWVPFRVKHSKQQK 0 0 ETPRMPLSDKSSVKEVFGIENLLNTSGSEHAQKPVCSPEEGNTSGQHFGKKL 1 2 KLRRKNVEVNRYRLRERKDLAYEEVSEPQDDDYL 1 2 YCEKCQNFFIDSCPNHGPPVFVKDSVVDRGHPNHSVLSLPPGLRIGPSGIPEAGLGVWNEASDLPVGLHFGPYKGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGQDESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGRELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 2 ELRTEIHPCFLCSLAFSSQKFLTQHVEWNHRTEIFPGASARINPKPGDPCPDQLQEHFDSQNKNDKASNEVKRKSKPRHKWTRQRISTAFSSTLKEQMRSEESKRTVEEELRTGQTTNIEDTAKSFIASETS RIERQCGQCFSDKSNVSEHQRTHTGEKP YICRECGRGFSQKSDLIKHQRTHTEEKP YICRECGRGFTQKSDLIKHQRTHTEEKP YICRECGRGFTQKSDLIKHQRTHTGEKP YICRECGRGFTQKSDLIKHQRTHTEEKP YICRECGRGFTQKSSLIRHQRTHTGEKP YICRECGLGFTQKSNLIRHLRTHTGEKP YICRECGLGFTRKSNLIQHQRTHTGEKP YICRECGQGLTWKSSLIQHQRTHTGEKP YICRECGRGFTWKSSLIQHQRTHTVEK >PRDM7_musMus Mus musculus (mouse) genomic strain C57BL/6J 12 repeats 12 PDCD2 chr17 CN723438 eight transcripts, four from retina 0 MNTNKLEENSPEEDTGKFEWKPK 0 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 2 GLRAPRPAFMCYQRQAMKPQINDSEDSDEEWTPKQQ 1 2 VSPPWVPFRVKHSKQQK 0 0 ESSRMPFSGESNVKEGSGIENLLNTSGSEHVQKPVSSLEEGNTSGQHSGKKL 1 2 KLRKKNVEVKMYRLRERKGLAYEEVSEPQDDDYL 1 2 YCEKCQNFFIDSCPNHGPPLFVKDSMVDRGHPNHSVLSLPPGLRISPSGIPEAGLGVWNEASDLPVGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGQDESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 2 ELRTEIHPCLLCSLAFSSQKFLTQHMEWNHRTEIFPGTSARINPKPGDPCSDQLQEQHVDSQNKNDKASNEVKRKSKPRQRISTTFPSTLKEQMRSEESKRTVEELRTGQTTNTEDTVKSFIASEIS SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQNSHLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREK >PRDM7_musMol Mus molossinus (wild_mouse) GU216230 Glir gene 11 noDet full length deposit 0 MNTNKLEENSPEEDTGKFEWKPK 0 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 2 GLRAPRPAFMCYQRQAMKPQINDSEDSDEEWTPKQQ 1 2 VSPPWVPFRVKHSKQQK 0 0 ESSRMPFSGESNVKEGSGIENLLNTSGSEHVQKPVSSLEEGNTSGQHSGKKL 1 2 KLRKKNVEVKMYRLRERKGLAYKEVSEPQDDDYL 1 2 YCEKCQNFFIDSCPNHGPPLFVKDSMVDRGHPNHSVLSLPPGLRISPSGIPEAGLGVWNEASDLPVGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGQDESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 2 ELRTEIHPCLLCSLAFSSQKFLTQHMEWNHRTEIFPGTSARINPKPGDPCSDQLQEQHVDSQNKNDKASNEVKRKSKPRQRISTTFPSTLKEQMRSEESKRTVEELRTGQTTNTEDTVKSFIASEIS SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSVLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTEKSSLIKHQRTHTGEKP YVCRECGWGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSSLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGWGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREK >PRDM7_criGri Cricetulus griseus (hamster) AFTD01086355 no out-of-frame continuation, no synteny information in contig, no PRDM9 0 MSCTRNTNKQEGNSPAGDAERLEWKPK 0 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 2 GLKAPRPAFMCYQRQAFKPQMDDSEDSDEEWTPKQQ 1 2 GSPPWVPFRVKHTKKQK 0 0 ETQRIPLNKESNVKEVSGSENLLSTSGSEHVQKTVFSPGEGNASGQHTGQKP 1 2 ELRRKNVEVKMYSLRERKDLAYEEVNEPQDDDYL 1 2 YCEKCQNFFINSCPSHGPPIFVKDSMVDRGHPNCSVLSLPPGLRIGPSGIPEAGLGVWNEASDLPVGLHFGPYKGQITDDEEAANSGYSWL 0 0 ITKGRNCYEYVDGQEESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVINPGCELLVWYGDEYGQELGIQWGRKNKKGFATGR 1 2 ELRTEIHPCLLCSLAFSSPKFLSQHVQWNHRTQIFPGASSTINSKPGDPHPDQLQEQQHFNSHNKNDKARSLEVKGKSKPMHKWTRQISTAFPSTLKGHMRSEENKKTMEVLRTGQKTNTEDTIKSFIGSEIS RIERKCGQYFSDKSNVNEHQRTHTGEKP YVCRECGRGFTQKSHLIRHQRTHTGEKP YVCRECGRGFTQKSNLIRHQRTHTGERP CVCLFKKDKKASVNKTTPQQSQKDKCSL* 0 >PRDM7_dipOrd Dipodomys ordii (kangaroo_rat) genome Glir gene -- noDet dubious fragment, no orthologous terminal exon 0 0 0 1 2 GLKAPRPVFMCHRRQAIKPQVDDTDDSDEEWTPGRQ 1 2 0 0 1 2 elRTKEVKMRMYSLRERKSYAYEEISEPQDDDYL 1 2 yCEQCQNFFINSCTVHGPPIFVRDNVVDKGHYDRSVLSLPPGLRIRQSSIPEAGLGVWNEESDLPLGLHFGPYEGQITEDEDAANSGYSWM 0 0 ITKGRNCYVYVDGKDKSQANWMR 2 1 YVNCARYDEEQNLVAFQYHRQIFYRTCRVIKAGCELLVWYGDEYGQELGIKWGSKWKRELTAgr 1 2 >PRDM7_speTri Spermophilus tridecemlineatus (squirrel) AAQQ01308561 Glir gene -- noDet plus exon by exon traces 0 0 0 1 2 GFRAPRPAFMCHQRQTIKLQMDDTEDSDEEWTPRQQ 1 2 0 0 LKPEVLLSNESSLKELSGTANLLNTSGSEQVQKPVSPLREASASRQHSRRKL 1 2 ELRTKEVEVKMYSLRERKGHAYKEVSEPQDDDYL 1 2 yCDKCQNFFMDSCPVHGPPTFIKDSVVNKDHSNHSTLSLPLGLRIGPSSIPEAGLGVWNEATDLPLGLHFGPYRGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELSAGR 1 2 EPKPEIHPCPSCSLAFSSQKFLSQHVDRSHPSQIFPGTSMRKKLIPGDSSPRDQLQEQQHPDPHGWNDKARGQEVQGSLKPTHKGTRQRGISSPPKGQMGRSEESERMMEDDLKADQEINPEDTDKILVGVEMSRI >PRDM7_bosTau Bos taurus (cow) pseudogene on chr18 main Gas8 URAH synteny block not qTer not old, splice sites preserved 0 MSPNRSPEESIEGDTGRTEWKPT 0 0 AKDAFKDISIYFCKEEWAQMG*WekA*YRNVKRNYEALITL 1 >PRDM7L1_bosTau Bos taurus (cattle) chrX:85801132 active gene 21.3 repeats formerly PRDM9b_bosTau, adjacent URAH pseudogene 0 MSPNRSPENSTEGDAGRTEWKPM 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIQYRNVKRNYEALIAI 1 2 GFRATQPGFMHHGRQVLKSQVDDTEDSDEEWTPRQQ 1 2 GKPSGMAFRGEPSKHPK 0 0 RLSRGPLNKVSSLKKLPGAAKLLKKSGSKQAQKPVPPPREARTPGKHPRHKV 1 2 ELRRKETEVKRYSVRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQIIYNEEDSNSGYCWL 0 0 VTKGRNSYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCRVVRPGCELLVWYGDEYGEELGIKQDKRGKSKLSAQR 1 2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNQQQRYSDPHSLSDKPEGREPKERPHPLLKGPKLCIRPKRISTASSYPPKGQMGGSEVHERMTEEPSTSQKLNPEDTGKLFMEAGVSGIVR VNYGDHEQGSKDRSSLITHEKIHTGEKP YVCKECGKSFNGRSDLTKHKRTHTGEKP YACGECGRSFSFKKNLITHKRTHTREKP YVCRECGRSFNEKSRLTIHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCGECGRSFNEKSRLTIHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCGECGQSFNEKSRLTIHKRTHTGEKP YACGDCGQSFSLKSVLITHQRTHTGEKP YVCGECGQSFNEKSRLTIHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCGDCGQSFSFKSVLITHQRTHTGEKP YACGECGRSFSGKSNLTKHKRTHTGEKP YVCGECGRSFSEKSNLTKHKRTHTGEKP YVCGDCGQSFSFKSVLITHQRTHTGEKP YVCGECGRSFSEKSNLTKHKRTHTGEKP YACGECGRSFSFKKNLITHQRTHTGEKP YVCRECGRSFSEKSRLTIHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCRECGRSFSVMSNLIRHQRTHTGEKP YVCRECGRSFRVKSNLVRHQRTHTGEKP YVCMECE* 0 >PRDM7L2_bosTau Bos taurus (cattle) appears intact KRAB SSXRD SET C2H2 0 MSPNRSPENSTEGDAGRTEWKPK 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATQPGFMHHRRQVLKPQVDDTEDSDEEWTPRQQ 1 2 GKPSGMAFRGERSKHQK 0 0 RLSRGPLNKVSSLKKLPGAAKLLKKSGSKQAQKPVPPPREARTPGKHPRHKV 1 2 ELRRKETKVKRYSVRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQIIYNEEDSHSGYCWL 0 0 VTKGRNSYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCRVVRPGCELLVWYGDEYGEELGIKQDKRGKSKLSAQR 1 2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNEQQRYSDPHSLSDKPEGREPKERPHPLLKGPKLCIRLKRISTASSYPPKGQMGGSEVHERMTEEPSTSQKLNPEDTGKLFMEAGVSGIVR VKYGEHEQDSKDKSSLITHEKIHTGEKP YVCTECGKSFNWKSDLTKHKRTHSEEKP YACGECGRSFSFKKNLIIHQRTHTGEKP YVCGECGRSFSEKSNLTKHKRTHTGEKP YACGECGQSFSFKKNLITHQRTHTGEKP YVCGECGRSFSEKSRLTTHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCRECGRSFSVISNLIRHQRTHTGEKP YVCRECEQSFREKSNLVRHQRTHTGEKP YVCMECE* 0 >PRDM7L3_bosTau Bos taurus (cattle) appears intact KRAB SSXRD SET C2H2 probable assembly stutter also has URAHps 0 MSPNRSPENSTEGDAGRTEWKPK 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATQPGFMHHRRQVLKPQVDDTEDSDEEWTPRQQ 1 2 GKPSGMAFRGERSKHQK 0 0 RLSRGPLNKVSSLKKLPGAAKLLKKSGSKQAQKPVPPPREARTPGKHPRHKV 1 2 ELRRKETKVKRYSVRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQIIYNEEDSHSGYCWL 0 0 VTKGRNSYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCRVVRPGCELLVWYGDEYGEELGIKQDKRGKSKLSAQR 1 2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNEQQRYSDPHSLSDKPEGREPKERPHPLLKGPKLCIRLKRISTASSYPPKGQMGGSEVHERMTEEPSTSQKLNPEDTGKLFMEAGVSGIVR VKYGEHEQDSKDKSSLITHEKIHTGEKP YVCTECGKSFNWKSDLTKHKRTHSEEKP YACGECGRSFSFKKNLIIHQRTHTGEKP YVCGECGRSFSEKSNLTKHKRTHTGEKP YACGECGQSFSFKKNLITHQRTHTGEKP YVCGECGRSFSEKSRLTTHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCRECGRSFSVISNLIRHQRTHTGEKP YVCRECEQSFREKSNLVRHQRTHTGEKP YVCMECE* 0 >PRDM7L4_bosTau Bos taurus (cattle) chr21 distal frameshift, stop codon, deletion: pseudogene adjacent to URAHL3 pseudogene 0 MNPYRSPEESTEGDAGRTEWRWR 0 0 1 2 1 2 GKPSWMTFRVKHSKHQK 0 0 gMSRALVSNKSSLKELPGASKLLKTSGPKQAQIQCPLPEKQVPLNSTLDKKW 1 2 GPIQKETEVKMYSLRERASHVYQEVSEPQDDDYL 1 2 YCEKCENF IKSCAVHGLPTFVKDCAVEKGHVNRLALSVPAGLSIRPSGIPEAVLGVWNEVSDLPLALHFGSYKGQIIDDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDTSWAKWMR 2 1 YVNCARDDKEQNLVAFLSHRQIFYQTCPVVRPGCELLVWYGDKYSQELSIKCGSRWKSELMASR 1 2 EPKPKIYPCASCSLAFSSQKFLSQHAEHNHPSQILLRTSARDRLQTKDSCPGNQNHQQQYSDPHSWRDKPEDREVKERPQPLLQSVRLRRVSRASSYSPKGQMGDTWVSERMMQEPSTGQKVNTEDTGKLCMGAGVLTIIR VKSveCGQDSKDRSSLITHQRIHTGEKP YACRECGRNFSEKSPLIRPQRTHIGEKP FVCRECE*GFSH IKHQRTHTGEKP YVCRECEQSFSEKSTLIRHQTTHTGEKP YVCGEVE* >PRDM7L5_bosTau Bos taurus (cattle) intact somewhat diverged chr1:161387010- no URAH 0 MRPNTSPEESTERDAGRTEWKPT 0 0 AKDAFKDISVYFSKEEWEEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATRPAFMHHRRQVIKLQADDTEDSDEEWTPRQQ 1 2 GKLSSMAFRVEHNKHQN 0 0 TMSRAPLSKEFSLKELPGAAKLLKTSGSKQAQKLVPPPGKARTPGQHPRQKV 1 2 ELRRKETEVKRYSLRERKGHVYQEVSEPQDDDYL 1 2 YCEECQDFFIDSCAAHGPPIFVKDCAVEKGHANRSALTLPPGLSIRESSIPEAGLGVWNEVSDLPLGLHFGPYEGQITDDEEAANSGYSWL 0 0 ITKRRNCYEYVDGKDTSLANWMR 2 1 RYVNCARDDEEQNLVALQYHGQIFYRTCQVVRPGCELLVWYGDEYGQDLGIKRESSRKSELAGPR 1 1 ESKPKIHPCASCSLAFSSQKFLSQHVQHNHPSQTLLRPSARDYLQPEDPCPGSQNQQQRYSDPHSPSDKPEGREVKDRPQPLLKSIRLKRISRASSYSPRGQMGASGVHERITEEPSTSQKPNPEDTGKLFMGAGVSGIIK VKYGECGQGSKDRSSLITNQRTHTGEKP YVCGECGQSFNQKSTLITHQRTHTGEKP YVCGECGRSFNQKSTLITHQRTHTGEKP YVCGECGRSFSQKSTLIKHQRTHTGEKP YVCGECGQSFNQKSTLITHQRTHTGEKP YVCGECGQSFNQKSTLITHQRTHTGEKP YVCGECGRSFSRKSTLITHQRTHRGEAL CLQGV* >PRDM7_oviAri Ovis aries (sheep) original PRDM7 pseudogene adjacent to GAS8 URAH on chr14, first two exons only (as in cow) 0 MSPNRFP*ESTGGDPGRTEWKPM 0 0 AKDAFKDISIYFSKEEWAEMR*W 1 >PRDM7L1_oviAri Ovis aries (sheep) active gene ChrX:5148004 frameshift exon 2 may be read error; translocated with URAH2 pseudogene to ChrX both minus strand 0 MSPNRSPEKSTEGDAGRTEWKPM 0 0 AKDAFKDISIYFTKEEWAEMGEWekVQYRNVKRNYKALIAI 1 2 GFRATQPAFMHHHRQVIKPQVDDTEDSEEEWTPRQQ 1 2 GKPSGMAFRGEPSKHQK 0 0 RLSRGPLNKVSSLKKLPGAAKLLKKTGSKQAQKPVPPPREARTPGQHPRHKV 1 2 ELRRKETEVKRYSLRERKGHVYQEVSELQDDDYL 1 2 yCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQVIYNEEASHSGYSWL 0 0 VTKGRNSYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCQVVRPGCELLVWYGDEYGEELGIKQDSRGKSKLSAQR 1 2 EPKPKIHPCASCSLSFSSQKFLSQHVQRSHPSQILLRPSPRDHLQPEDPCPGKQNQQQRYSDPHSPSDKPEGQEPKERPHPLLKGPKLCIRLKRISTASSYTPKGQMGGSEVHEKMTEEPSTSQKLNPENTGKLFMEAGVSGIVR VKYGEHEQGSKDKSSLITHERIHTGEKP YVCKECGKSFNGRSNLTRHKRTHTGEKP YVCRECGQSFSLKSILITHQRTHTGEKP YVCGECGQSFSEKSNLTRHKRTHTGEKP YVCRECGQSFSLKSILITHQRTHTGEKP YVCRECGRSFSVKSNLTRHKMTHTGEKP YVCGECGQSFSQKPHLIKHQRTHTGEKP YVCRECGRSFSAMSNLIRHQRTHTGEKP YVCRECGRSFSAMSNLIRHQRTHTGEKP YVCRECGRQVSSHTRGHTQGRSPMFAGSVGEASV* >PRDM7L2_oviAri Ovis aries (sheep) OARX:67135299 not tandem, two frameshifts and internal stop: pseudogene 0 0 0 1 2 1 2 0 0 GMSRGPLSKVSSLKKLPGTTKLLKTSGSKQAQKPVPSSREARTSG*HTRQKV 1 2 eLRRKETEVKRYSL*ERKGHVYQVVSEPQNDNYL 1 67134368 2 ycEECQNFFMNSCAAQVPPTFVKDSAVGKGHANCSALTLPPGLSIRLSGIPEARLGVWNEVSDLPLGLHFGpyEGQITDDEEAAHSGDSWL 0 0 ITKGRNSFEYVDGKDMSLANWMR 2 67133322 1 CVKCTQDNKEQNLVALQYHRQIFYRICQVVRPGcqLLVWYGDEYAVELGIKWDNRGKSEFTARR 2 ELKPKIHPCASCSPAFSSQKFLSQYVQPNHPSQILLRPSARDHLQPEDPCPGNQNEQQ*YSDPHSPSDKPEGCKAKERPPWLLKSMSVRISMASSYSPKGQMRGSETHYRMTEEPSTSQKLNPEDIGKLFMGTGVSGIIK IKYEECGQVSKDRSSLITHEGTHTREQS YVCRECGQSFSVKSSLIRLQRTHTGEKP YT ESVGKASVRTPISSHTRGHTQERSP MVAGRVGKASVRTQFSSDNRGHTQGRSP MFSGSVGKASVRSPSSSHTRGHAPE >PRDM7L3_oviAri Ovis aries (sheep) chrX:67,417,035+ 2 internal stops: pseudogene 0 MS*NRSPQERTEGDAGRTEWKLM 0 0 ANGAFKNISIYFSKEEWAEMGEWEKI*YGNVKRNCEALIAI 1 2 GFRATRPAFMHHRRQVIKPQGNDTEDSDEEWTPWQQ 1 2 0 0 GKSRGPLSKASSLKKLPGAAKLLKKSGSKWAQEPVKPPRETRTPGQHSRQKV 1 2 ELGRKETDMKRYSLRERKGHVYQEVSEPQDDDYL 1 2 yCQECQNFFINSCDAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRLSGIPEAGLGVWNEASHLPLGLHFGPYEGQITDDKEAVNSGYSWL 0 0 ITKGRKSYEYVDGKDTSLANWMR 2 1 YVNCARHYEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGEKLGIRCESRGKSMLAAGR 1 2 * 0 >PRDM7L4_oviAri Ovis aries (sheep) OAR18:28239884-one frameshift, one stop codon pseudogene paired with URAH3 pseudogene 0 MNPYISPEESTEGDAERTEWKPM 0 0 AKDAFKDISIYFSKEECAEMGEWGKICYRNAKRNCEALITI 1 2 GLRATRLAFMHHCRQVIKPQVDDIEDSDEEWTPRQQ 1 2 0 0 EGMSKALVSNKSSLKEMPGASKLLKTRGPKQAQIPVPAPREPSTSEQHPRQKV 1 2 1 2 CCHGLPTLVKDCAVEKGHANHSALSLSPGSSIRPSGIPEAGLGVWNKVSDLLLGLHFGSYVGQITDDEEAAKSGYSWL 0 0 ITKGRNCYEYVDGKQRSWANWIr 2 1 YVNGAQD KEQNLVAFLTHRQIFY*TCRVVRPGCELLVWYRDTYSQELSIKCGSRWKSELTASR 1 2 EPKPKIHPCVSCSLsfSSQKFLRQHVERNHPSQILLRTSARDRLQTKDSCPGNQNHEQQYCDPHSWSDKPEDGEVRERPQPLLKSIRLRRVSRASSYSPKGQMGDSWVSEKMMEEPSTGQKLNTEGTGKLCMGAGVLRIIR VKHGECGQGSKDRSSLITHQRIHNGEKP YVCRECGQSFSEKSILIRHQRTHTGEKP FVCRECERGFSQKSYLIRHQKTHTGEKS YVCREVE* >PRDM7L5_oviAri Ovis aries (sheep) OAR5:40765355+ 1 frameshift and 4 internal stops: pseudogene 0 MSSNRSLKERTEGDARRTEWKPMV 0 0 AKDAFKDISI*FSKEEWAEMGE*EKI*YRNVKRNYEALITTI 1 2 GLRAP*PPFMYHRRQVIKPQVDDIEDSDEEWTPRQQ 1 2 0 0 GMSRVPLSN ESMKELLGAAKLLT SGSKQAQKPVPPPREASTSEQHPRKKV 1 2 ELRRKETEMKIYSLQKRKGHMYQEVSDPQDDNYL 1 2 0 2 ycEKCQNF INSCAAHGPPTFVKDCVVEKGHASCSALtlSPGLSIRPSGIPEAGLRVWNEASDLPLGLHFGPYKGQITDDEEVANSRYFWL 0 0 2 0 ITKGRNCYEYVDGRD 1 2 1 YVNCAQDDEEQNLVAFQYHRQIFS*TCWVVRPGCELLVWYRDEYGQELSIK*GSRHKSELTVRR 1 2 PMCSCSLAFSSQKFLSQHVKCNHPSQILLKTSARDRLQPEDPCPGNPNQQQQYSDLHSWSDKPESRESKEKPQPLLKS----IRLRRISRASSYSSRGQMGGFRVHKRMREEPSTGKEVSPEDAGKLFMGEGVSRIM VKYGDCG*GSKDRSSLMTHQRTHTGENP >PRDM9d_munMun Muntiacus muntjak (muntjac) AC216498 Laur gene 4 noDet frameshift exon 9 no syntenic loci; identities: 92%b 89%a 90%c 0 MRPNRSQEESTEGNAGRTERKPT 0 0 GKDAFKDISVYFSKEEWEEMGEWEKIRYRNMKRNYEALIAI 1 2 GFRATQPTFMHHRRQVIKSQVDDTEDSDEEWTPRQQ 1 2 GKPSSMAFRVEHSKNQK 0 0 RMSRAPLSNESGLKELPGAAKSLKTSDSKQARNPVPHHRKARTPGQLPRQKV 1 2 ELRRKETGVKRYSLRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFINSCAAHGPpTFVKDCAVEKGHANRSALTLPHGLSIRLSGIPDAGLGVWNKVSDLALGLHFGPYKGQITDNEEAANSGYAWL 0 0 ITKGRNCYEYVDGKDTSWANWMR 2 1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGQDFGIKRNSRGKSELAAGR 1 2 EPKPKIHPCASCSLTFSSQKFLSQHIQCSHPPQTLLRPSERDLLQPEDPCPGNQNQQQRYSDPHSPSDKPEGHEAKDRPQPLLKSIRLKRISRASSCSPRGQMGGSGVHERMTEEPSTSQKLNPGDTGTLLTGAGVSGIMK VKYGECGQGSKDRSSLSTHERTHTGEKP YVCRECGQSFSGKPVLIRHQRTHTGEKP YVCMECGRSFSAKSVLMTHHRTHTGEKP YICRECGQSFSQKIHLIRHQRIHTGE.P SVFRECE >PRDM9c_munMun Muntiacus muntjak (muntjac) AC154919 Laur gene 15 noDet no syntenic loci AC204173 99% identical 0 MRPNRSPEESTEGDAGRTEQKPT 0 0 AKDAFKDISVYFSKEEWEEMGDWEKIRYRNMKRNYEVLIAI 1 2 GFRATRPDFMHHRRQVIKPQVDDTEDSDEEWAPRQQ 1 2 GKPSSVAFRVEHSKHQK 0 0 RMSRAPLSNESGLKELPGAAKPLKTSGSKQAQNPVPHHRKARTPGQLPRQKV 1 2 ELRRKETGVKRYSLRERKGHVYQEVSKPQDDDYL 1 2 YCEKCQNFFIDSCAAHGPPTFVKDCAVEKGHANRSLLTLPPGLSIRLSGIPDAGLGVWNEASDLPLGLHFGPYEGQITDDEEAANSGYAWL 0 0 ITKGRDCYQYVDGKDTSWANWMR 2 1 YVNCARDDEEQNLVAFQYHGQIFYQTCQVVRPGCELLVWCGDEYGQDLGIKRNSRGKSELVAGR 1 2 EPKPKIHPCASCSLAFSSQKFLSQHIQRSHPSQTLLRPSERDLLQPEDPCPGNQNQRFSDPHRPSDRPQPLLKSIRLKRISRASSYSPRGQMGGSGVHELMTEEPSTSHKLNPEDTGTLLMGAGVSGIMR VTYGECGQGSKDRSSLTTHERTYTGEKP YVCGECGRSFCQKAHLITHQRTHTGEKP YVCRECGQSFSRNSLLIRHQRIHTGEKP YVCGECGRSFRDKSNLISHRRTHTGEKP YVCGECGQSFSDKSNLIRHQRTHAGEKP YVCGECGRSFNRKSHLITHQRTHTGEKP YACRECGQSFSQKSILITHQRTHTGEKP YACRECG.SFSQKSILITHQRTHTGEKP YVCGECGRSFSQKSLLITHQRTHTGEKP YVCMECGRSFSQKTHLITHQRTHTGEKP YVCGECGRSFSQKSLLITHQRTHTGEKP YVCGECGRSFSQKSLLITHQRTHTGEKP YICMECGRSFSQKTHLITHQRTHTGEKP YVCGKCGQSFSDKSNLISHKRTHTGEKP YVCRECGRSFNRKSLLITHQRTHT.E.P YVCRECE >PRDM9b_munMun Muntiacus muntjak (muntjac) AC218859 Laur gene 13 noDet no syntenic loci 0 MRPNTSPEESTEGDAGRTERKPT 0 0 AKDAFKDISVYFSKEEWEEMGDWEKSRYRNMKRNYEVLIAI 1 2 GFRATRPDFMHHRRQVIKPQVDDTEDSDEEWAPRQQ 1 2 GKPSSMAFRVEHSKHQK 0 0 RMSRAPLSNESGLKELPGAAKPLKTSGSKQAQNPVPHHRKARTPGQLPRQKV 1 2 ELRRKETGVKRYSLRERKGHVYQEVSKPQDDDYL 1 2 YCEECQNFFIDSCAAHGPPTFVKDCAVEKGHANRSALTLPPGLSIRLSGIPDAGLGVWNETSDLPLGLHFGPYEGQITDDEEAANSGYAWL 0 0 ITKGRNCYQYVDGKDTSWANWMR 2 1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVIRPGCELLVWYGDEYGQDLGIKRNSRGKSELATGR 1 2 EPKPKIHPCASCSLAFSSQKFLSQHIQRSHPSQTLLRPSERDLLQPEDPCPGSQNQRYSDPHSPSDKPEGQEAKDRPQQLLKSIRLKRISRASSYSPGGQMGGSGVHERMTEEPSTSQKLNPEDTGTLLTGAGVSGIMR VTYGECWKGSKDRSSLTTHERTHTGEKP YVCGECGQSFHHGSVLIRHQRTHTGEKP YVCGECGRSFSQKSVLIRHQRTHTGEKP YVCGECGRSFSQKSVLIRHQRTHTGEKP YVCGECGRSFSQKAHLITHQRTHTGEKP YVCGECGRSFSQKTHLISHKRTHTGEKP YVCGECGRSFCQKSALIRHQRAHTGEKP YVCGECGRSFIQKSDFIRHQRTHTGEKP YVCRECGQSYSDKTVLITHERTHTGEKP YVCGECGRSYSDKTVLITHERTHTGEKP YVCGECGRSFLWKSALIRHQRTHTGEKP YACGDCGRSFNQKSNFIRHQRTHTGEKP YVCGECWRSFSQKSSSSDTRGHTQGRRP VCRECG SFSQKSHLISHQRTHTEEKP YVCRECE >PRDM9a_munMun Muntiacus muntjak (muntjac) AC225653 Laur gene 7 noDet unordered contigs htgs; no synteny tag stop instead of aag K 0 MRPNRSPEESTEGDAGRTEQKPT 0 0 AKDAFKDISVYFSKEEWEEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATRPDFMHHCRQVIKPQVDDTEDSDEEWTPRQQ 1 2 GKPSSMAFRVKHSKHQK 0 0 GMSRAPLIKESSLKELLGAAKLMKTSGSKQAQNPVPHPRKARTPGQHPRQKV 1 2 ELTRKETGVKRYSLRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFIDSCAAHGLPTFVKDCAVEKGHANRSALTLPPGLSIRLSGIPDAGLGVWNEESDLPLGLHFGPYEGQITDDEEAANSGYAWL 0 0 ITKGRNCYQYVDGKDTSWANWMR 2 1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVIRPGCELLVWYGDEYGQDLGIKRNSRGKSELAAGR 1 2 EPKPKIHPCASCSLAFTSQKFLSQHIQRSHPAQTLLRPSERNLLQPEHPCPGSQNQRYSDPHSLSDKPEGQEAKDRPQPLLKSIRLKRISRASSYSPGGQMGGSGVHERMKDEPSTSQKLNPEDTGTLLTGAGVSGIMR VTYGECGKGSKDRSSLTTHERTHTGEKP YACRECGRSFRQKSDFITHQRTHTGEKP YVCGQCGRSFGRKFALIRHQRIHTGEKP YVCRECGQSFSQKTHLSSHQRTHTGEKP YVCGECGRSFSQKSVLIRHQRTHTGEKP YVCQECGRSFSDKSNLISHKRTHMGEKP YVCRECGRSFIRKSVLIRHQRTHTGE.P YVCRECE >PRDM9a_odoVir Odocoileus virginianus (deer) AEGZ01043838/AEGZ01024932/AEGZ01044038/AEGY01012861/AEGZ01038568/AEGZ01003977/AEGY01011331/AEGZ01039403/AEGY01006151 possibly chimeric, frameshifts possibly errors in low coverage assembly 0 MTVSRSLEESTGGDAGRTEWKPT 0 0 AFKDISKYFSKEEWARLGYSEKISYVYMKRNYETMTRL 1 2 GFRATRPAFMHHRRQVIKPQVDDTEDSDEEWTPRQQ 1 2 GKPSRMALREEHIKHQK 0 0 RMSRAPLSKESSLKELPGAAKSLKTSGSKQAQKPVPHPRKARTPGQHPRQKV 1 2 ELRRKETGVKRYSLRERKGHVYQEVSEPQDDDYL 1 2 yCEECQNFFIDSCAAHGPPTFVKDSVVKRGHANRSALTLPPGLSIRLSGIPDAGLGVWNEASDLPRGLHFGPYEGQITDDEEAANSGYAWL 0 0 ITKGRNCYEYVDGKDTSWANWMR 2 1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGQFLGIKRDSRgKSKLAAGR 1 2 EPKPKIHPCASCSLAFSSQKFLSQHIQCSHPSQTPPrPSERDLLQPEDPCPGNQNQRYSDPHSPSDKPEGQEAKDRPQPLLKSIRLKRISRASSYSPGGQMGGSGVHE >PRDM7_bosTau Bos taurus (cattle) genome Laur pseu -- GAS8+ missing C2H2 0 MSPNRSPEESIEGDTGRTEWKPT 0 0 AKDAFKDISIYFCKEEWAQMG WEKIRYRNVKRNYEALITL 1 2 1 2 0 0 1 2 1 2 0 0 2 1 1 2 >PRDM7_turTru Tursiops truncatus (dolphin) ABRN01441536 Laur gene 9 gas8+ no useful synteny 0 MSTDRWPEDSTEGDAGRTAWKPT 0 0 VKDAFKDISIYFSKEEWTEMGEWEKIRYRNVKKNYEALVTL 1 2 GLRAPRPAFMCHRRQAIKAQVGDPEDSDEEWTPRQQ 1 2 VKPSWVAFRVEHSKHQK 0 0 AVPPVPLSNESSLKKLPGAAQLQKASGPAQAQSPAPPPGAASTSAWHTRQKL 1 2 ERRAKQIEVKMYSLRERKGHVYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAAHGAPTFVKDSAVEKGHPNRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDTSWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYSQELGIPWGSGWKSQLVaGR 1 2 DPKPKIQPCGSCSLAFSSQKILSQHVECSHPSQVLPRTSARDRVQPEDPCPGYQNRQQQYSDPHSWSNKPECQEVKERSKPLLKRIRLGRISRAFSSSPKGQMGSSRAHERMMEAGPSTGQKVNPEATGKLLIGAGVSRVVK VKYRSSGQGSKDRSSLTKHQRTHTGEKP YVCGECGRDFSLKSDLIRHQRTHTGEKP YVCGECGRDFSLKSGLISHQRTHTGEKP YVCGECGRDFSQKSGLIRHQRTHTGEKP YVCGECGRDFSLKSGLISHQRTHTGEKP YVCGECGRDFSQKSGLIRHQRTHTGEKP YVCGECGRDFSLKSGLITHQRTHTGEKP YVCGECGRDFSQKSNLITHQRTHTGEKP YVCGECGRDFSRKSSYI >PRDM7_susScr Sus scrofa (pig) FP476134 Laur gene 9 GAS8+ unordered HTGS not wgs misassembly or inversion; not in genome browser 0 MRPDRRPEESPDPAAGSTERKAA 0 0 ATDAFKDISIYFSKEEWTEMGEWEKIRYRNVKRNYEALTTI 1 2 GLRAPRPAFMCHRRQAIKPQVDDTEDSDEEWTPRQQ 1 2 VKPCRVAFRVEHNKHQK 0 0 SDSRVPLSNKSSLKELLTTAEVPETSGSEQAQEPVSPPGEASTSRRRSGQEL 1 2 ARRRKDTEARMYSLRERKGHAYQEVGEPQDDDYL 1 2 yCEKCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALTLPPGLRIRPSGIPEAGLGVWNEAHDLPLGLHFGPYEGQVTEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGI 1 2 EPKPKIHPCPSCSLAFSSQRFLSQHVERSHPSQSLPRASARRGLQPEGPCPDNQQQQQPYPDPHSWDGTSESQDVKEGSKPFLERRRLRKTSRASSYAPEGQMRSSRVRERMTEEEPSAGQKVNPEDTGTLFTVAGES GILRVENRGYGPDSGLTRHPRTHTGEKP HVCSECGRGFSVKSHLIRHQRTHTGEKP YVCRECGRGFSVKSHLIRHQRTHTGEKP YVCRECGRGFSVKSSLITHQRTHTGEKP YVCRECGRGFSVKSHLIRHQRTHTGEKP YVCRECGRGFSEKSSLVTHQRTHTGEKP FVCRECGRGFSVKSSLVTHQRTHTGEKP YVCRECGRGFSVKSNFITHQRTHTGEKP YVCRECGRGFSEKSSLVTHQRTHTGEKP YVCREGE >PRDM7_lamPac Lama pacos (llama) scaffolds traces 0 0 0 TFKDISIYFSKEEWTEMGEWEKIRYRNVKRNYEALITI 1 2 GLRAPRPAFMCHRRKAIKPQVDDTEDSDEEWTPRQQ 1 2 0 0 GMPRGPLSNQSSLKELSGTAKPLKTSGSGQAQKPFPPLGEASTSGRHSRQKL 1 2 ELRRKESQVKMYSLRERKGHAYQEVSEPQDDDYL 1 2 0 0 ITKGRKCYEYVDGKDKYWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGEEYGQELGIKWGSKWKKSLWQGE 1 2 EPKIYLCPSCSLAFSSQKFLSQHVKHNHPSQILPRTAAGRHLEPEDPCPGNQNEQQQHSDQHSWNDKPEGQEAKERSKPFLKRIRLRRISGAFSYSHKGQMGNSRVHDRMIEEEPSTGQKVNPKDTGKLFTWAGVSRTVE VNYGEYGQGCKDTSHLTTHQRTHTGEKP YVCRECGRGFTRKSNLTIHQREHTTGEK >PRDM7_canFam Canis familiaris (dog) genome Laur pseu 5 GAS8+ frameshift fixed to 6 ZNF; synteny MNS1 K1F1B intervening CDH3 oddity 0 0 0 AFKDISKYFSKEEWAKLGYSDKITYVYMKRNYDTMTGL 1 2 GLRATLPAFMCPKKRAIKSK RHDSDENENHRNQ 1 2 VKPSWVAFRMEQSKHQK 0 0 GIPRVPLSNKSSLKELSETAKLLNTSSPEQGQKSVSLPGKASTSGHHTRQKL 1 2 ELRRKDVEVKMYSLQERKGLAYQEVSEPQDDDYL 1 2 yCEK*QTFFIDSCTVHGPPTFVKDSEVDKGQPNHSALTLPPGLRIRTSSIPQAGLGVWN*ASDLPLGLHFGPYKGQITEDEEAANSGYSCL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YMNCARDDEEQsLVAFQYHRQIFYRtPGHQASCELLVWYGDEYSQELGIKWGSKWKSELTAGK 1 2 EPNPEIHPCPSCSLaFSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECGrGFIQRSNLSIHQRTHTGEKP YVCRECGRGFTQRSTLNEHQRTHTEEKP YVCRECGRSFTRRSTLITHQRTHTGEKP YVCRECGRSFT >PRDM7_canLup Canis lupus (gray_wolf) JF750654 first frameshift: 1 bp del ggcctt to gcctt; second frameshift 1 bp del ggaca to ggacGa ePNPEIHPCPSCSLaFSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECGrGFIQRSNLSIHQRTHTGEKP YVCRECGRGFTQRSTLNEHQRTHTEEKP YVCRECGRSFTRRSTLITHQRTHTGEKP YVCRECGRSFTKRS >PRDM7_canAur Canis aureus (golden_jackal) JF750659 EPNPEIHPCPSCSLaFSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECGRGFTQRSTLNEHQRTHTEEKP YVCRECGRSFTKRST >PRDM7_lycPic Lycaon pictus (painted_dog) F750657 EPNPEIHPCPSCSLaFSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWSDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIMr VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRGFTHRTNLIIHQRTHTGEKP YVCRECGrGFIQRSNLSIHQRTHTGEKP YVCRECGRGFTQRSTLNEHQRTHTEEKP YVCRECGRSFTRRSTLITHQRTHTGEKP YVCRECGRSFTKRST >PRDM7_canMes Canis mesomelas (black-backed_jackal) JF750658 EPNPEIHPCPSCSLaFSSQKFLSQHLEHNHPSQILPQISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRDFTHRTNLIIHQRTHTGEKP YVCRECGrGFIQRSNLSIHQRTHTGEKP YVCRECGRGFTQRSTLNEHQRTHTEEKP YVCRECGRSFtRRSTLITHQRTHTGEKP YVCRECGRSFTKRST >PRDM7_speVen Speothos venaticus (bush_dog) JF750656 EPNPEIHPCPSCSLaFSSQKFLSQHLEHNHPSQILP*ISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKRIRQRRISRAFSTPCKGQTTCEGIVKEEPSASSQKLNPEDTGKLFKGVGMTRIIR VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECgRGFTHRTNLIIHQTTHTGEKP YVCRECGrGFIQRSNLSIHQRTHTGEKP YVCRECGRSFT*RSTFSThQRTH >PRDM7_vulVul Vulpes vulpes (red_fox) JF750655 more distal frameshif EPNPEIYPCPSCSLSFSSQKFLSQHLEHNHPSQILPRISIREHFQPKDPCPGCQNQQQQQHSDPQCWNDRAKGQEGKERFKPLPkSIRQRKISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR VKYRGCGRGFNDRSHLSRHQRTHMGENP YVCRECGRGFTHRTNLIIHQRTHTGEKP YVSWECGRSFTRRSNLITHQRTHTGEKP YVCRECGRGFTKRSTLSTHQRTHL >PRDM7_neoVis Neovison vison (mink) JF288183 anomalous array, contig terminates 0 0 0 1 2 1 2 VRPSWVAFRMEQSKHQK 0 0 GIPRAPLSNESSLKELSETAKLLNTSGSEQGQKPVSHPGEASTSGHHSLRKL 1 2 ELRRKDVEVKMYSLRERKSLAYQEVSEPQDDDYL 1 2 YCEKCQNFFIDSCAVHGPPTFVKDSAVDKGQPNRSDLTLPPGLRIRPSGIPQAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 ITKGRNCYEYVDGKDNSWANWMR 2 1 1 2 EPKPEIHPCPSCTLAFSSQKFLSQHLKCNHPSQILPRISAGEHFQPEDPCPGEQNHQQQQHSDPQSWNDKAKGQEVKESFKPLLESIRQRRNSRAFPTPCKGQTGYEGMVEEESSTGQKLNPEETEKLFMGVGMSRMIR VKYRGSGQGFDDRSHLSRHQRTHKEEKP SVGKELRREFIHKSVLVTHQRTHTEALP >PRDM7_musPut Mustela putorius (ferret) AEYP01035076 AEYP01035077 AEYP01035078:GAS8- HUIH+ CAD1L+ distal PRDM7+ 0 MRPRTASESEQGLPGGPSTGSVSGPPEETPERDSGRTGRKPP 0 0 AQDAFKDISVYFSKEEWTEMGDWEKIRYRNVKRNYEALITI 1 2 GLRAPRPAFMCHRRQATIPRVDDTEDSDEEWTPRQQ 1 2 VRPSWVAFKMEQSKHQK 0 0 GVPRAPLSNESSLKELSETAKLLNTSGSEHDQKPVSHPGEASTSGHHSLRKL 1 2 ELRRKDVEVKMYSLRERKSLAYQEVSEPQDDDYL 1 2 YCEKCQNFFIDSCAVHGPPTFVKDSAVDKGQPNRSALTLPPGLRIRPSGIPQAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDNSWANWMR 2 1 YVNCARDDEEQNLVAFQYRRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKSELTAEK 1 2 EPKPEIHPCPSCTLAFSSQKFLSQHLERNHPSQILPRISAGEHFQPEDPCPGEQNHQQQQHSDPQNWNDKAKGQDVKESFKPLLESIRQRKNSRAFPIPCEGQTGYEGIVEEEPSTGQKLNPEETGKLFMGVGMSRIIR VKYRGSGQGFDDRSHLSRHQRTHKEEKP SVGKEPRREFIHKSVLVTHQRTHTGEKP YVCRECGRGFTQRSHLIRHQRthtgEKP YVCRECGRGFTQRSNLITHHRTHTGEKP YVCRECGRGFTRRSNLIRHHRTHTGEKP YVCRECGRGFTWRSHLITHQRTHTGEKP YVCRECGRGFTWRSHLIRHQRTHTGEKP YVCRECGRGFTRRSNLITHQRTHTEALP INCISMTRGKM* >PRDM7_ailMel Ailuropoda melanoleuca (panda) GL193502 Laur gene 6 GAS8+ first three exons from different contig ACTA01106867 0 MGPLPASESEQSLPGGPSTMSLNTSPEETPERDSGRTGWKPT 0 0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYEALITI 1 2 GLRAPRPAFMCHRRQAIKPQVDDTEDSDEEWTPRRQ 1 2 VRPSWVAFRMEQSKHQR 0 0 GIPRAPLRNESSLKELSETAKLLNTSGSELGQKPVSLPGEASTSGHDSLQKL 1 2 GFRRKDVEVKMYSLRERKSLAYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAVHGPPTFVKDSAVDKGQPNRSALTLPPGLRIRPSGIPQAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDNSWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKSELAAGK 1 2 EPKPEIHPCPSCSLAFSSQKFLSQHLEHNHPSQILSRKSASEHFQQEDPCPGHQNQQQQQHSDPHRWNDKAKGQEVKERFKPLLKSIRQRRISRAFSSPCKGQTRSSTVCEGMVEEEPSAGQKLNPEETGKLFMGVGMSGIIR VKYRGCGRDFSDRSHQSGHQRRH QKKP SVCKKVKREFSHKSVLITHQRTHTGEKP YVCRECGRGFTQRSNLIRHQRTHTGEKP YVCRECGRGFTQRSNLIRHQRTHTGEKP YVCRECGRGFTQRSSLIRHQRTHTGEKP YVCRECGRGFTLRPNLIGHQRTHTEALP INYISTTKEQM >PRDM7_felCat Felis catus (cat) genome Laur gene 11 GAS8+ two contigs GAS8 implied by downstream CAD1 0 MEPSPASESARGQPGGPGTTSPLRFPEQSAERGSRKARWKPT 0 0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYEALMTI 1 2 gLRAPRPAFMCHRRQAIKPQVDVTEDSDEEWTPRQQ 1 2 VKPSWVASRVDQNKQHK 0 0 GTHRVPLSKESSLKDFSETAKLLNTSGSEQGQKPVSLPGEASTSGHHSRRKL 1 2 ELRRKEIGVKMYSLRERKGFAYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAVHGPPTFVKDNAVGKGHPNRSALTLPPGLRIRPSSIPEAGLGVWNEASDLPLGTHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDNSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKSELSTGK 1 2 EPQPDIHRCPSCSLAFSSQKFLSQHVECKHSSQSLPQISARKHFQPENPCPGDQNQQQQQHSDPHSWNDKAKCQEVKERSRPLLKSIKQRRISRAFSTPCKGQMGSSRVCEGMVEEGPSMGQNLNSEDTGKLFMGVGMSRIVR IKNRGCEQGFNDRSHFSRHQRTHKEEKP SVCNEFRRDFSHKSALITHQRTHTGEKP YVCRECGRGFTQRSNLFRHQRTHTGEKP YVCRECGRGFTQRSDLFTHQRTHTGEKP YVCRECGRGFTRRSNLFTHQRTHTGEKP YVCRECGRGFTRRSHLFTHQRTHTGEKP YVCRECGRGFTQRSNLFTHQRTHTGEKP YVCRECGRGFTQRSDLFRHQRTHTGEKP YVCRECGRGFTQRSHLFTHQRTHTGEKP YVCRECGRGFTQRSNLFRHQRTHTGEKP YVCRECGRGFTWRSNLFTHQRTHTGEKP YVCRKDGQGFTNKLHLSYQRT NVATTHSIPQL >PRDM7_equCab Equus caballus (horse) genome Laur gene 4 GAS8+ missing front exons, pre-terminal stop GAS8+- flanked right by EMR2- 0 0 0 1 2 1 2 VKPSWVAFRVEQSKQQK 0 0 RMRTAPLSNESRLKELSGTAKLLKTSSSEQVQKPVSPLGEASSSEQHSRRKL 1 2 ELRRKEVGVKMYSLRERKGHAYQEVSEPQDDDYL 1 2 yCENCQNFFIDSCAAHGPPIFVKDSAVDKGHPNRSALTLPLGLRIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDISWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYGQELGIKWGSKWKRELTAGR 1 2 EPKLEIHPCPSCSLAFSSQKFLSQHVERNHPSQILPGTSARNHLQPEDPSPGDQNQQQQHSDPHSWKDKAHSQEVKERSKPLLKKIRQRRIPRAFSYPPKGQMENFRMRERIMEEKPSIGRKVNPEDTGKLFLEMRMSRNVR VQYGGCGRGFNDRASLIKHQRTHTGEKP YVCRECEQGFTQKSSLIAHQRTHTGEKP YVCRECEQGFSEKSHLIRHQRTHTGEKP YVCRECEQGFSVKSNLIRHQRTHTGEKL *FCREGK >PRDM7_pteVam Pteropus vampyrus (bat) ABRP01250178 Laur gene 7 GAS8+ 4 distal exons of GAS8+-; unique F sweep in zinc finger; 15 ZNF dotplot no CAD1 0 MRPDRSPEEAPEGDTRRTGCKPK 0 0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYDALQAI 1 2 GLRAPRPAFMCRRRQAIKPQVDDSEDSDEEWTPRQQ 1 2 0 0 AMPRVPLSNEPSLKELSVIANLLKASGSEQDQKPVFPPGKASASRQHSRQKL 1 2 GLRRKGVEVKMYSLRERTGRVYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAAHGSPIFVKDSEVDIRHPNRSALTLPPGLRIGPSGIPEAGLGVWNEASDLPLGLLFGPYEGQVTEDEEAANSGYSWL 0 0 QGKGRNCYEYVDGKDESRANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKRELTAGR 1 2 EPKPAIHPCPSCSLAFSGQKFLSQHMKRSHPSQSLPGISARKHLQSKEPHPEDQSQQQHNDPRSWNDKAEGQEVKERSKPLLERNRQRKIFRAFSKPPKGQMGSPREYERMMEAEPSTSQKVNPENTGKSSVGVGASRIVI VKYGGCEHGFDDGSHLIMHQRTHSGEKP FVCRECERGFSKKSNLITHQRTHSGEKP FVCRECERGFTRKSSLITHQRTHSGEKP FVCRECERGFTQKSHLITHQRTHSGEKP FVCRECERGFSEKSSLIKHQRTHSGEKP FVCRECERGFTRKSSLITHQRTHSGEKP FVCRECERGFTQKSSLIKHQRTHSGEKP FVCRECERGFTQKSSLIKHQRTHSGEKP FVCRECERGFTQKSSLIKHQRTHSGEKP FVCRECERGFTQKSSLITHQRTHSGEKP FVCRECERGFTQKSHLITHQRTHSGEKP FVCRECERGFSKKSNLITHQRTHSGEKP FVCRECERGFTRKSLLITHQRTHSGEKP FVFRECERGFTQKSSLITHQRTHSGEKP FVCRECERGFTRKSYLITHQRTHSGEKP FVGRECE >PRDM9_pteVam Pteropus vampyrus (bat) ABRP01232219 Laur pseu 15 noDet frameshift ttt to tttt fixed in last zinc finger; no blastx synteny 0 0 0 1 2 1 2 vQPSWVAFGVEQSKHQK 0 0 AMPRVPLSNESSLKELSVIANPLKASGSEQNQQPVFPPGKASASRQHSRRKL 1 2 eLRRKGVEVKMDSLRERMGRVYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAAHGSPIFVKDSEVDIGHPNHSALTLPPGLRIGPSGIPEAGLGVWNEASNLPLGLLFGPYEGQVTEDEEAANSKYSwM 0 0 spKGETAEYV DGKDESRANWMR 2 1 YVNCARDDEDQNLVAFQFRRQIFYRTCRVIMPGCELLVWYGDEYGQGLGIKWGSKWKREFTAGR 1 2 EPKPEIHPCPSCSLAFSSRKFLSQHMKRSHPSQSLPGISARKHLQSKEPHPEDQSQQQQQQQHTDPCSWNDKAEGQEVKERSKPMLERNGQRKISRAFSKPPKGQMGSPRECERMMEAEPSTSQKVNPENTGKSSVGVGASRIVR VKYGGCGHGFDDGSHFIRHQRTHSGEKP FVCRECERGFNEKSSLTMHQRTHSGEKP FVCREC.EGFSVKSSLIRHQRTYSGEKP FVCRECEQGFNEKSSLTMHQRTHSGEKP FFCRECEGFSVK.SSLIRHQRTHSGQKP FVCRECKRGFTQKSHLITHQRTHSGEKP FCRECER.GFTQKSHLIKHQRTHSGEKP FVCRECA >PRDM7_myoLuc Myotis lucifugus (bat) AAPE02062260 Laur gene 6 gas8+ TGA stop codon; CpG hotspot for R CGA; SXXRD implies missing KRAB no CAD1 0 0 0 1 2 1 2 0 0 AKSRAPLSNESSLKELSGTANLLTTSGSEQTQKTVPPPGEASTSGQHPRSKL 1 2 dLRRKEIEVKMYSLRERKCRVYQEISEPQDDDYL 1 2 YCEKCQNFFIDSCAVHGPPTFVKDSAVDKGHANRSALTLPPGLRIGPSGIPEAGLGVWNEECDLPVGLHYGPYEGQITEDEAIANSGYSWL 0 0 ITKGRNCYEYVDGKDTSQANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVVRKGCELLVWYGEEYGQELGIKWGSKWKTEPVAGR 1 2 EPKPEIHPCPSCSVAFSSQTFLSQHGKRNHPSEILPGAPAGNHLQSEEPGPERQNQQQQQQTGPHGWNDKAEGQEVKGRSKPLLKRIRQRGTSRASFKPPNRHMGSSSERERIREEEPSTGQNVNHKNTGKLFVGVKRSKSVT IKHGGCGQGFNDGSHIDTHQRTHSGEKP YICRECGGFTHKSDL IRHQRTHSQENP YVCRECGRGFRDRSTLITHQRTHSGEKP YVCRECGRGLTEKSTLITHQRTHSGEKP YVCRECGRGFTRKSTLITHQRTHSGEKP YVCRECGRGSRVKSNLIRHQRTHSGEKS GVCIEGE >PRDM7_sorAra Sorex araneus (shrew) AALT01000095 Laur gene 8 noDet no useful synteny; upstream spectrin, IgG; GAS8 contig has no sign of pseudogene 0 MSLNRPAEMNTQGKARKLMLKPM 0 0 SKDAFKDISMYFSKEEWAEMGDWEKIRHRNVKRNYEELISI 1 2 GLRAARPAFMSHRRQAIKTQLDDTEESDEEWTPNQQ 1 2 VKSLRVAFRAEQSKHQK 0 0 GRSRTPISNESSSKELSGTRTLLNTKCTKQAQKPLFPPGEASTSGHYSKPKL 1 2 ELRRKEPEVKMYSLRERKGRAYQEVSEPQDDDYL 1 2 YCENCQNFFINKCSAHGSPIFVKDNAVAKGHSNRSALTLPHGLRIGPSGIPEAGLGIWNEASDLPLGLHFGPYEGQITNDEEAANSGYSWL 0 0 ITKGRNCYEYVDGVDESLANWMR 2 1 YVNCARDYEEQNLVAFQYHRQIFYRTCRIIKPGCELLVWYGDEYGQELGIKWGSKWKSELTADK 1 2 EPKPEIYPCPCCSLAFSNQKFLSRHVEHSHPSLILPGTSARTHPKSVNFCPGDQNQWQQHSDACNDKPDEPWNDKLENHKSKGRSKPLPKRMGQKRISTAFPNLRSSKMGSSNKHETIMDKINTGQKENPKDTYRVFAGIGMPRIIR DKHVTLRRSFTNRSSPLTHQRTHTGEKP YVCRECGRGFSQKSHLLTHQRTHTGEKP YVCRECGRGFTDRSSLLTHQRTHTGEKP YVCRECGRGFSLKSSLLRHQRTHTGEKP YVCRECGRGFSLKSSLLTHQRTHTGEKP YVCRECGRGFTDRSSLLTHQRTHTGEKP YVCRECGRGFSLKSSLLTHQRTHTGEKP YVCRECGRGFSRKSSLLRHQRTHTGEKP YVCES >PRDM7_echEur Echinops europaeus (hedgehog) ti|970966337 epkpeihpcPSCSLAFSAQKFLNQHVKHSHPSQILPGTSTRKQPQVENPCLSNQNQQKQHSNFQNQHDSTESQEAIEKFKPLLKMIKQKTISNGFSKLPKEQIGSSREHEKTKEEESNSCQKMNPEDTSELLVGLGMSRIV DKYEGSGKNYYDMSHIITHQKTHTGEKP HVCKECGRGFSEKSSLIAHQRTHTGEKP HVCRECGRGFSEKSSLIAHQRAHTGEKP HVCRECGRGFSEKSGLITHQSTHTGEKP HVCRECGrGFSAKSSLIAHKRTHTGEKA PCLQGSVGELQ >PRDM9a_loxAfr Loxodonta africana (elephant) genome Afro gene 12 noDet chr 153 novel synteny THEG+ MIER2+ PPAP2C PRDM9- ZNF699- 0 MSPARAAKKNPRGDVGSAGRTPT 0 0 aKDTFRDISIYFSKEEWAEMGEWEKFRYRNVKRNYEALVTI 1 2 GLRAPRPAFMCHRRQAIKAQVDNTEDSDEEWTPRQQ 1 2 VKPPSVASRAEQSRHQK 0 0 GTPKALLGNESSLKEVSGTAILLNTTGSEQAQKPVSSPGEASTSDQPSRWKL 1 2 EPRRNEVEVKMYNLRERKGLEYQEVSEPQDDDYL 1 2 yCEKCQNFFIDTCAVHGAPMFVKDSPVDRGHPNHSALTLPPGLRIGPSSIPKAGLGVWNEASELPLGLHFGPYEGQVTEDKEAANSGYSWL 0 0 ITKGKNCYEYVDGKDESWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRTIQPDCELLVWYGDEYGQELGIKWGSRWKKELTSGR 1 2 EPKPEIHPCPSCRLAFSSQKFLSQHMKHSHPSPPFPGTPERKYLQPEDPRPGGRRQQRSEQHMWSDKAEDPEAGDGSRLVFERTRRGCISKACSSLPKGQIGSSREGNRMMETKPSPGQKANPEDAEKLFLGVGTSRIAK VRCGECGQGFSQKSVLIRHQKTHSGEKP YVCGECGRGFSVKSVLIKHQRTHSGEKP YVCGECGRGFSVKSVLITHQRTHSGEKP YVCGECGRGFSVKSVLITHQRTHSGEKP YVCGECGRGFSQKSDLIKHQRTHSGEKP YSCRECGRGFSRKSVLITHQRTHSGEKP YVCGECGRGFSQKSNLITHQRTHSGEKP YVCGECGRGFSRKSVLITHQRTHSGEKP YVCGECGRGFSQKSNLITHQRTHSGEKP YVCGECGRGFSQKSDLITHQRTHSGEKP YVCRECGRGFSRKSNLITHQRTHSGEKP YVCRECRRGFSVKSALI GHGRRKCSKSAEPLHFPRVSRDQK >PRDM9b_loxAfr Loxodonta africana (elephant) genome Afro pseu 3 noDet approx seq after frameshift correction 0 0 0 1 2 1 2 0 0 GTPKVLLSNESSLKEVSGTAILLSTMGSEQAQKPVSSPGEASTSDQPSRRKQ 1 2 EPRRKEVEVNMYSLRERKGLVYQEVGEPQDDDYL 1 2 yCEKCQNFFIHTCAVHGAPMFVKDSHVDRGHLNHSALTLPPGLRIGPSSIPEAGLRVR EVSEQLLGLHIGPYEGQVTEDkEAAHSGYSWL 0 0 ITKGRNCYKYVDGKDDPWANRMR 2 1 YVNCIQD KEQNLVAFQYHRQIFHWTCCTIRPGCELLVWYGDNYSQELGIKWGSR KKEL 1 2 EPKPEIHPCPSCPLAISSQKFLDQHTKHSHPSPPFPGTPERKHLQPEDPHPGGRRQQHSEQHLNDKAEDPETGDGSKPVFERARLVGGGAGGVSKVCSSLPKGQMGSSREGNRMMETEGQKVNPEDTEKLFLGVGISRLAK VRCGEYGQGFSQKSVLIRHQRTYSGEEH YVCGECGRGFSWKSQLTRHQRSHSWEKP YVCRECGGFSVKSTLI GTGEGNAATIHLHLPS >PRDM7_loxAfr Loxodonta africana (elephant) genome Afro pseu 5 GAS8+ scaffold_57 several frameshifts; ZNF540 opposite strand upstream of N-terminus 0 0 0 1 2 GLRASHPAFTCHCMQAIKAQMDDTEDSNEEQTPRQq 1 2 VRPSWVAFRMEQSKHQR 0 0 GMLRVPRSNESSLKNLSGTSIMLSRAGSEQAQKLVLPPGKASTSDEHSRQKP 1 2 EHRRKGVEVKMYSF ERKGLVYQEIS PQDDDYL 1 2 YCEKCQNFFIDTCESHGVPTFVKNSTTDSGHPNHLALTPSSGLRTRPSSIPKAWLRLWNKAFELLLGLPFSPCEGQVIEDEAVDNSGYSWL 0 0 2 1 YVNGTQDEKEQNLVFFQYHRQIFYQTCYAVWPGCQLLVWYRDECGQELGIKWDNRGKKEFe 1 2 EPKPEAHPCPSCPLAFSSEKFLSQHMKHNHPSQSSPETPERKHLQPEDPHPGHQNQQQQQHSDPHRWNDKAEGQQTGDRSKPMFENIRQEVTSRAFSSLPKGQMVCSREGNRMMETEPSPGLKVNPEVTGKLFLGVESSRIAK VKYRGCGRDFSDRSHQSGHQRRHQ KKP SVCKKVKREFSHKSVLITHQRTHSGEKS YVCKESGRGFSAKSNLIRPRRTHTGEKP YVCGERGG.FSVSGLII.HQRAHSPEKP YVCREGRRGFGDKSSFIKHQRATLGEKS YVCKESGRGFS AKSNLIRPRRKKCRHDTTPHPQL >PRDM9_triMan Trichechus manatus (manatee) AHIN01064530 synteny: none (12740 bp) contig terminates prior to repeat 0 MSPARATEESPGGDARRTPT 0 0 AKDAFRDISIYFSKEEWAEMGEWEKFRYRNVKRNYEALVAI 1 2 GLRAPRPAFMCHRRQAIKAQVDDTEDSDEEWTPRQQ 1 2 VKPPWVVSRVEQSKHQK 0 0 GTPRAPLNNESSLKEVSGTEILLSTAGSEQAQKLVSSPGEASTSDQHSRQKL 1 2 EPRRKEVEVKMYSLRERKGLAYQEVAEPQDDDYL 1 2 yCEKCQNFFIDACAVHGAPTFVKDSPADRGHPNRSALTLPPGLGIGPSGIPKAGLGVWNEASELPLGVHFGPYEGQVTEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDESWANWMR 2 1 YVNCARNEEEQNLVAFQYHRQIFYRTCSTIQPGCELLVWYGDEYGQELGIKWGSRWKKELTSGR 1 2 EPKPEIHPCPSCPLAFSSQKFLSQHVKHRHPSQPFSGTPARKHLQPEDPRPGDQRQQHSERTQNDKAEDRETGDGSKRVFERTREGETSKVDSSLPKGQIGSSREGNRMMETEPSPGQKVNPEDTEKLLLGVGISRIVK VRHGECGQGFSQKSVLITHQRTHSGEKP YVCRECGRGFS >PRDM7_triMan Trichechus manatus (manatee) pseudogene AHIN01061278 internal stop and frameshift 0 0 0 AKDAFRDISIYFSKEGWEEMGEWEKFRYRNMKRN^VQRNYKALVTI 1 2 GLKVPHPAFMCH*RQSIKSQTDDTEDSHEEWASRQQ 1 2 0 0 GILRASLSNKSSLKELSGT-IMLSRAGPEQAQKSVLPPGEASTSDKHSRQKL 1 2 EPRRKEVEVKTYNL*ERKDLVYQEVS*PQDGDYL 1 2 yCEKCQNF-TDSCAAHGDPTFVKDSAMDSGHPHHS------GLGIGPSSIPKARLEVWNKA GLHFSPYEGQVTEEEEAANSSYSWV 0 0 2 1 YVNYTQDKE*QNLVAFQYHRQIFYRTCRAIWPGCELLVWYGDEYGQELDIKWNSR-QKEFTAGR 1 >PRDM7_echTel Echinops telfairi (tenrec) genome Afro pseu 5 noDet 2 frameshifts plus stop codon 0 0 0 1 2 GLRAPRPAFMCHHRPAAKGQVEDSEDSDEEWTPRQR 1 2 0 0 GMPGVSLRNESNLKVLSGTAILLTAAEPEQPH PGSPPGEATTSHEHLRQKV 1 2 epELRRRAVMMNSLRERKNLMYQEVSTPCDDNCL 1 2 YGERCHNFFIDTHIAHGATTFVKDS PMDRSNCSILPPGLRIGPSGIPEAGLGVWNEASELPLGLHFVPYEGQVTKDEAATNSGYSWM 0 0 ITKGRNCYEYVDGKDKSWANwMr 2 1 1 2 EPKPEVNPCPSCPLALSSQQLKHSHPFQSLPGTPAEKHLQAEDFHPRGQKLHHFEHHIRNERAEGLETGDGSKPMLERTRLGKMSKTTYNSPKGQTRSSGETNRIREADLNPGQGVNAEDTRNLFLGIGISRIAK VRCRECGHGFSVKSSLITHQRIHTGEKP YVCSECGQGFSQKSVLIRHQRIHTGEKP YICRECDRGFSRKSHLIKHQRTHSGEKP YVCRECGQGFSQKSVLITHHRTHSGEKP YVCRECGRGFSQKSDLIKHERTHS >PRDM7a_proCap Procavia capensis (hyrax) ABRQ01227339 Afro pseu 17 noDet frameshift and two stop codons in exon 10 0 0 0 AKDAFRDISIYFSKEEWAEMGEWEKSRYRNVKRNYEALVAI 1 2 GLRAPRPAFMCHRRQAIKAQVDNTEDSDEEWTPRQQ 1 2 AKPRSVASREELRKPQK 0 0 GTPKALLGNESSLKEVSGTAILLNTTGSEQAQKPVSSPGEASTSDQPSRWKL 1 2 EPRRKEAEVKRYNLREGTNPAYQEVGDTQDDDYL 1 2 yCEKYQKFCTDVCPAHGALAFLKDLSVERGHPKHSALTLPPGLRIGASGIPEAGLGVWSEASELPPGLHFGPCERQVTKDNEAANRGYLWP 0 0 ITKGRSCSLYMDRKDESRANWMR 2 1 YVRHAGDKEEQNLVAFQYHRQIFYRTCRPVQPGCELLVWPGAEDGQELGLQRGSRWKKELASQT 1 2 EARPEIHPCPSCPLAFSTPKFLSHHVKHSHPCQPFPGTLARRPLQPEDPHPGDRRQQHSEQPNWNDKAEGPEIGHVSRPVFEKTRQEGFSEARSSLPKGQMGRSREAERTTETQNSPGQKVNPEDTEILFLRGGISEIAK VKCGECGQGFSRKSHLIRHQRTHSGMKP YVCRECRRGFGVKSLLTRHQRTCSGMKP YVCRECGQGFRWKSHLIRHQRTHSGEKP FVCSECGRGFSVRSHLFTHQRTHSGEKP YVCKECGRGFSVKSYLTTHQRTHTGEKP YVCKECGRGFSWKSHLITHQRTHSGEKP YVCRQCGRGFSVQSHLIIHQRTHSGDKP YICRECGRDFTEKSSLIRHRRTHSGEKP YVCRDCG*GFTRKSLLITHQRTHSGEKP YVYRECGRGFSCKSYLISHQKTHLGEKP YVCSDCGRGFSVKSQLVSHKRTHSGEKP FVCREC*RGFSVKSSLISHQRTHSGEKP FVCRECGRGFSVKSSLIKHQRTHSGEKP YVCKECGRGFSQKSSLITHQRTHSGEKP YVCRECGRGFGLKSYLITHQRTHTGEKP YICRECG*GFSVKSSLITDQRTHTGEKP YVCRECGRAFSKKSSLISHHRTHPAEAV YVHRECG >PRDM7b_proCap Procavia capensis (hyrax) ABRQ01392668 Afro pseu 13 noDet CpG stop in ZNF1, 4aa insert exon 4, frameshift exon 5 c to cc, 4aa del exon 9 etc 0 0 0 AKEYFRDISMFFS*ERWVEMSESEKFCYRNMKRNCETTGAG 1 2 GIRVFHPAFMIHPRKTIKAQMDDSEDSDEDWTARQQ 1 2 AKPPSVASREELRKPQK 0 0 GPSRAPLRIKSSLKRVSEPAIVWSTADSEQAQERVQKPVLSRREASASDQPLRRKV 1 2 EPRRHEAEDKRYSLRGGTGPACQEVGEPQDDDYL 1 2 yCEECRNFFIDTCVAHGTPVFIKDISVERGHPNRLALTLPTGLRIGPSSIPDAGLGVWNEASELPPGLHFGPCEGQVTEDEEAANSGYSWL 0 0 VTKGRSCFEYVDGKNEALANWMR 2 1 YVRRARDTEERNLVAFQYHRQIFYRTCCTVRPGCELLVWRGAEDSQALGSRRTMELTSQK 1 2 EARPEIHPCPSCPLAFSTQKFLSYHVNHSHSSEPFPGTHARRHLPREDPRPGYERDQRSEQHNWNDSTGGPERDVSRP VIERTWEGEISEACSSLPRGHMGRSREGERMAETQSSPGLKVTLAK VRWDEYGQGFGPKSHHITQQTKHSGKKP CVCKECG*GFRVKSLLKSHQMTHSGEKP YVCRECGRGFSVKSTLITHQRTHSGEKP YVCRECGRGFSVKSFLISHQRTHSGEKP YVCRECGRGFSWKSGLITHQRTHTGEKR YVCRECGHGFNRPSRLIRHQRTHSGEQP YVCRECGHGFNRRSQLIRHQRTHTGEQP YVCRECGQGFSGKSGLNRHQRTHSGEKP YVYKECGRGFSVKSTLIKHQRGHSGEKP YVCKECGRGFSRNSGLITHQRTHSGEQP YVCRECGRGFNQKSGVISHQRIHSGEKP FVCGECGRRFSWQSNLITHQRTHSGEKP FVCRECGRGFSAKTSLINHQRIH*GKKP YVCRDGG >PRDM7_dasNov Dasypus novemcinctus (armadillo) AAGV020462211 9 xena pseu TRAPP 0 0 0 AQDAFRDISTYFSREEWAEMGRWEKLRYRNVKRNYEALLAI 1 2 GLRAPRPAFMCHRKQSIKPQVDDAEDSDEEWTPRQQ 1 2 0 0 1 2 EPRRKGIDVKMYSLRERKGLAYEEVSEPQDDDYL 1 2 yCEKCQNFFIDSCTVHGPPIFVKDSAVDKGHPNRSALTLPSGLRIGPSGIPEAGLGIWNEASDLPLGLHFGPYEGQVTEDEEAANSGYSWL 0 0 ITKGRNYYEYEDGKDKSWANWMR 2 1 YVNCAWDDKEQNLVAFQYHRQIFYRTCRTIRPGCELLVWYGDEYGQELGIKWGSKWKKEFMTGT 1 2 ELKPEIHPCPSCPLAFSSEKFLSQHVRRHHPSQSFPAACAREHFQPQNPRPRGEEQQQHSDQCGWKDKAEGQETENRPKPLFERIKPMGSPRAFYNPPRGQMRSSREGKRMMEIQPSQDQKMNSE RGQLFLGVGIFKTEV IKFGENRQDFSDKSDHTSHQRTHTGEKP YVCRECGRGFSNNSHLTRHQRTHTGVKP YVCRECGQGFSVKPALTKHQRTHTVEKP yVCSECG GFSVKSTLITHQRTHTGEKP CVCRECGRGFNNKPDLTKHQRTHTGEKS YVCRECG GFSVKSTLIIHQRTHTGEKP YVCRECGRGFSEKSNLTVHQRTHTGEKP YVCRECGRGFSEKSNLTVHQRTHTGEKP YVCRECGRSFSVKSTLITHQRTHTVEKP YVCMKSEVVVSNKSHLNSHRRMKCGHRT PPPPQL >PRDM7_choHof Choloepus hoffmanni (sloth) ABVD01893961 2 xena gene noDet 0 0 0 1 2 1 2 0 0 1 2 1 2 ycekcQNFFFENCAAHGPPTLLKDSAVGQGRPKHSALVLPPGLRLGPSGIPEAGLGVWNEASDLPLGLHFGPYEGQVTEDEEATNSGYSWL 0 0 ITKGRNCYEYVDGKDKSCANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRAIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAEK 1 2 GLKPEIHPCPSCPLAFSTEKFLSQHVQRNHPSQIFPVTYARKHLQPQDPRPGDQQQPQPHSDQCHCSDKAEDQETEKRSKPLFESTKQMGISRAYSSPPEGQMRSSREDKRTMEIEPSQDQKMNPEETRLFVGVGILKTAR IKCGEYGQGFSVKPNLTTHQRTHTEEKP YVCRECGRGFGQKPNLSRHQRTHTGEKP YVCRECGRGFG >PRDMx_monDom Monodelphis domestica (opossum) gene genome no GAS8 fragment KRAB SSXRD SET weak C2H2 domain 0 0 0 GEDAFKDISTYFSKKQWVKLKEWEKVRLKNVKRNYEAMIKI 1 2 GLSVPRPAFMCRGRQNKKVKVEESGDSDEEWIPKQL 1 2 VKTLRFPSRAKQRTHPK 0 0 1 2 DCRRKDVEVHIYSLRERKYQVYQEMWDPQDDDYL 1 2 yCEECQIFFLDSCPLHGPPTFVQDSAMVKGHPYCSAITLPPGLRIGLSGIPGAGLGVWNEASTLPLGLHFGPYKGKMTEDDEAANSGYSWM 0 0 ITKGRNCYEYVDGKEESCSNWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKRPLPELTGE 1 2 GKPGISLCPSTLWASPLIPSSINTRCSKQPP*VFLDSGTGKL*AGRSTAGPATSNRFQLLSDKETSPKEHPSSLWGKTKQVDRREKFSLPQSQQVRGKESSSGEDLSRIQGKSTRQTTMAFQERNR KECE*GFTHQTNLVTHRWTHSGERP YVCV*GFTQKLGFSPYTWTL >PRDMx_macEug Macropus eugenii (wallaby) ABQO010244377 ABQO010410412 ABQO011136158 ABQO010410657 0 0 0 GEDTYKDISMYFSKKQWMELREWEKIRLKNVKQNYEAMIKI 1 2 gFSAPRPTFMCHGKQNKEAKVEESGDFDEEWIRKQP 1 2 0 0 1 2 ECRRKEAEVHIYNLRERKYQVYQEIWDPQDDDYL 1 2 FCEECQTFFLETCAVHGPPKFVQDSVMVKGHPYCSAITLPPGLRIGLSGIPGAGLGIWNEASNLPLGLHFGPYEGQMTEDDEAANSGYSWM 0 0 2 1 YVNCARDEEEQNLVAFQYHRKIFYRTCQIIRPGCELLVWYGDEYGQELGIKWGSKWKRPPITLT 1 2 espGIHVCPFCPLGSPLMHSQSTYAAQTSPQICLDSRTRNNYEPDQLLPPSSSCVSDKVEISQKQRPSSLCGKTKQVNLVEMLSLPQSPQVSKKSSSMDWDVSRIQGKSAKQTTQGFQKGDKKGFGS YKCGEYKQGFTSKSVLNRHRQKHSGKKP YVCEECGRGFTQVSNLTTHRQTHSGEKP YVCEECGRGFARKLNLTTHRRTHSGEKP YVCEECGRGFTQGSSLITHRRTHSGEKP YVCEECGRGFAWKLNLTTHRRTHSGEKP YVCKECGRGFTQGSSLITHRRTHSGEKP YVCKECGRGFTQGSNLTTHRRTHSGEKP YVCKECGRGFAWKSNLTTHRRTHSGEKP YVCKECGRGFTQVSNLIAHRRTHSGEKF YVYGQE FTWKSDLSTCR >PRDMx_sarHar Sarcophilus harrisii (tasmanian_devil) AFEY01386448 distal frameshifts and stop codons, syntenic -PSMC4 0 0 0 EEDSFKDISMYFSKKQWMELRDWEKVRFKNVKRNYEAMIKI 1 2 GLTASRPTFMCRGKQNRRAKVEESGDSDEEWMPKQL 1 2 VKASRFSSRLKQKTHLR 0 0 1 2 eCRKKDAAVHIYNLRERKYPIYQEIWDPQDDDYL 1 2 FCEECQTFFLETCAVHGPPKFVQDGAMIKGYPYCSAITLPPGLRIGLSGIPNAGLGVWNEGSNLPMGLHFGPYEGKSTEDDEAANSGYSWM 0 0 ITKGRNSYEYVDGKEESCSNWMR 2 1 YVNCAREEEEQNLVAFQYQRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGRKWKRPLTGIT 1 2 GIHLCPSCPSDFSTHAFLSQQVPKQPSQGFLDSTTGSHGLGNLHPDQLLPPGYSCVSDKAETSRKEHPSTLWEKIKKVDLEEPASLPQRQHVREEESNLGEWDLSRIQGESVKLTSLALQEESQEGLGQ YKCGEGKQRYSSKPGLIRHRQRTHSGEKC YVCEECKRGFARRSYLNIHRRRHSGQKP HVCEECKRGFADKSTLIRHRWTHSKEKP YICEECKQGFTQKSYLIKHRWKHLGEKP YVCKECKQRFTQRSYLNTHRWRHRQRSL vFCaECR*GFT*RS*LIIHRWTHSGERP YVCEECKGGFTQRSYLNtHTDGNVGKEEP YVCEECR >PRDMxa_ornAna Ornithorhynchus anatinus (platypus) gene genome chrX5 fragment X5 +- 20577549 no iMet possible in first exon phase 2 0 0 0 1 2 1 2 0 0 1 2 RIGKKPQVRDFNLRKQKRKIYNENYRPEDDDYL 1 2 yCEICQTFFLEKCVLHGPPVFVQDLPVEKWRPNRSTITLPPGMQIKVSGIPNAGLGVWNQATSLPRGLHFGPYMGIRTKNEKESHSGYSWM 0 2 IVRGKNYEYLDGKDKAFSNWMR 2 1 YVNCARSEREQNLVAIQYQGEIYYRTCRVIPPGQELLVWYGLEYGRHLGILPNNNNPEP 1 2 ERAKARVRKSERIEKAMARVRKSEQIERAKARVRTSERIERAMATV RKSERIERAKVTVKKSEQIERAMGRVRKSERIERAKDMGRKKALGGLPRPCRGGLSDETQQRKGGGHEQLGQKPGPSEA RAGPAEGSATPRR HCCDVCRKAFKRLSHLRQHKRIHTGEKP LVCKVCRRTFSDPSNLNRHSRIHTGLRP YVCKLCRKAFADPSNLKRHVFSHTGHKP FVCEKCGKGFNRCDNLKDHSAKHSEDNSTPKP >PRDMxb_ornAna Ornithorhynchus anatinus (platypus) gene genome chrX5 tandem fragment slight frameshift taa to ta YVN exon X5 +- 20605294 20611704 no iMet possible in first exon phase 2 gg as expected 0 0 0 1 2 1 2 0 0 1 2 RSGKKPQVRDFNLRKQKRKMYTEESEPEDDDYL 1 2 yCEDCQTFFLEKCSVHGPPVFVQDCEAKRCQQNRSEVTLPPGLLIKMSGIPNAGLGVWNQATSLPRGLYFGPFVGIRKNNVKDSLSGYSWA 0 0 ILRGRNYEYLDGKNTSFSNWMR 2 1 YVNCPRTKYEQNLVAIQYHREIYYRTTPCDSTRSRVAGVVWRRVRSYLGIFWKSETPKS 1 2 ERPHSSGGSFAPSARSGGVKQRIWSKRRSAALQRTRERRNSTHDFPPKHEDTAARQDERQCPDRGRAKQRGVRKSEQIERAKAMGRKKALGGLSPPRRERLSDEAGQRKKSGHEQFWQKPGPSEAWAGPAEGSTIPRR HCCDVCGKAFNRLSRLKQHKRVHTGEKP LVCKICKRAFSDPSNLNRHAKRHTGEKP FVCRVCGRSFNRSDNMNEHRWKHTSNNIIP NTGHMSATVVENASLCINRNYQIYKERATYL
>PRDMx_danRer Danio rerio (zebrafish) Q6P2A1 transcript BC064665 no KRAB SSXRD or exon 5 but knuckle SET early ZNf C2H2 array 0 MSLSP 1 2 DLPPSEEQNLEIQGSATNCYSVVIIEEQDDTFNDQPF 1 2 YCEMCQQHFIDQCETHGPPSFTCDSPAALGTPQRALLTLPQGLVIGRSSISHAGLGVFNQGQTVPLGMHFGPFDGEEISEEKALDSANSWV 0 0 ICRGNNQYSYIDAEKDTHSNWMK 2 1 FVVCSRSETEQNLVAFQQNGRILFRCCRPISPGQEFRVWYAEEYAQGLGAIWDKIWDNKCISQ 1 2 GSTEEQATQNCPCPFCHYSFPTLVYLHAHVKRTHPNEYAQFTQTHPLESEAHTPITEVEQCLVASDEALSTQTQPVTESPQEQISTQNGQPIHQTENSDEPDASDIYTAAGEISDEI HACVDCGRSFLRSCHLKRHQRTIHSKEKP YCCSQCKKCFSQATGLKRHQHTHQEQEKNIESPDRPSDI YPCTKCTLSFVAKINLHQHLKRHHHGEYLRLVESGSLTAETEEDHT EVCFDKQDPNYEPPSRGRKSTKNSLKGRGCPKKVAVGRPRGRPPKNKNLEVEVQKIS PICTNCEQSFSDLETLKTHQCPRRDDEGDNVEHPQEASQ YICGECIRAFSNLDLLKAHECIQQGEGS YCCPHCDLYFNRMCNLRRHERTIHSKEKP YCCTVCLKSFTQSSGLKRHQQSHLRRKSHRQSSALFTAAI FPCAYCPFSFTDERYLYKHIRRHHPEMSLKYLSFQEGGVLSVEKP HSCSQCCKSFSTIKGFKNHSCFKQGEKV YLCPDCGKAFSWFNSLKQHQRIHTGEKP YTCSQCGKSFVHSGQLNVHLRTHTGEKP FLCSQCGESFRQSGDLRRHEQKHSGVRP CQCPDCGKSFSRPQSLKAHQQLHVGTKL FPCTQCGKSFTRRYHLTRHHQKMHS >PRDMx_salSal Salmo salar NM_001173912 0 MESEWKSGGEEESGSEGERTPSSSHRDP 1 2 VCVSEQMKRAWLRQMNLRSRARVGYTEEEELRDEEYF 1 2 FCEECKSFFIEECELHGPPLFIPDTPAPLGAPDRARLTLPPGLEVRTSAIPGAGLGVFNHGHSVTQGTHYGPYEGELTDKELDMESGYSWV 0 0 IYKSKQRDEYIDGKRDTHSNWMR 2 1 YVNCARSEDEQNLVAFQYRGGILYRCCKPIAVGEELLVWYGEKYARDLGIVFDFLWDKKCSAR 1 2 GVNESSQSQIFSCSGCLFSFTAQTYLYKHIKRCHREECVRLPRSGGIRAETLAPPSGSQRCSTTPDRTPITLLTQKHRDTGKPAP HHCSQCGKSFRRSGDLKVHQRTHTGERP YHCSQCGKRFSVSGHLKTHQRTHTGERP YHCSQCGKSFCRSGDLKVHQRTHTGERP YHCSQCGKRFSVSRHLKRHQHIHTGERP YHCSQCGKSFSASWSVKRHQITVHSVGRVSVSQEA >PRDMx_oncMyk Oncorhynchus mykiss testis FP324541 CR372724 0 mTPSSSHRDPVC 1 2 VSEQRKRAWLKQVNLCSRARVRVGYTEEEELREEDYF 1 2 FCEECKSFFIEECELHGPPLFIQDTPAPLGAPDRARLTLPPGLEVRTSAIPGAGLGVFNYGHSVTQGTHYGPYEGELTDTELAMESGYSWV 0 0 IYKSKQSDEYIDAKRETHSNWMR 2 1 YVNCARNEEEQNLVAFQYRGGILYRCCKPLAVGEELLVWYGEEYARDLGIIFDFLWDRKSSAR 1 2 GVNESSQSQIFSCSGCPFSFTAQIYLYKHTKRCHREEYVRLPRSGGIRSETLAPPSGSQRCSTTPDRTPITLLTQKHQDTGKPRP HHCSQCGKSFHRSGDLKVHQRTHTGERP YHCSQCGKRFSVSGNLKTHQRIHTGERP YPCSQCGKSFHRSDLKVHQRTHTGEKP YHCSQCGKRFSVSGNLKTHQRIHTGERL YPCSQCGKSFHRSELKVQQRTRPGKKTISLFPVWE >PRDMx_ictPun Ictalurus punctatus FD367165 FD063496 C-terminus missing second gene present 0 MKTEAKDGGTEGI 2 1 VKKETLELSISNHGNSFHIIPEVVSIKEEEADVKDFL 1 2 YCEVCKSVFFSKCEVHGPALFIADSPVPMGVADRARQTLPPGLEIQKSGIPDAGLGVFNKGETVPVGAHFGPYQGELVDKEEAMNSVYSWV 0 0 IYMSRQCEKYIDAKREVHANWMR 2 1 YVNCAHSDGEQNLVAFQYRGGILYRCCRPINPGQELLVWYEEKYASDVGPIFAQLWNIKCSLSGKVHT
Other genes of relevance
It is instructive to consider certain closely related placental KRAB, ZNF and PRDM genes that may have some connection to the origin of PRDM7 and PRDM9. Nomenclature is very unsatisfactory in these gene families, as can be seen from lack of correspondence between gene name and intronation which is exceedingly well conserved in metazoa. For example, HKR1 a conventional ZNF family member, is egregiously misnamed. The methylase component is exceedingly old with clear antecedents in bacteria. Evidently gene duplications in an early intronless stem eukaryote were subsequently intronated randomly in different paralogs and shuffled into various larger proteins. Within PRDM*, the gene tree is (((PRDM7/9,PRDM11),(PRDM4,PRDM10)),PRDM6) with others only related by a PR (SET) domain. PRDM11 and its novel giant terminal exon are discussed in a separate article
A set of fragmentary sequences from murid rodents is also of some comparative interest. These include common strain variants of lab mouse as well as close relatives. Only the terminal zinc finger array is available for most of these. While these are likely PRDM7 (rather than PRDM9 which rodents never had), it is not possible to decisively establish this with GAS8 synteny in any of the rodents (or lagomorphs) currently the subject of a genome project.
>PRDM11_homSap Homo sapiens (human) corrected 511+722 aa PhosS 3RAY coverage knuckle SET ZnF_TTF hATC_dimerization no early zinc finger or terminal array syn PFM8 related ZNF 862 0 MTENMKECLAQTNAAVGDMVTVVKTEVCSPLRDQEYGQPC 2 1 SRRPDSSAMEVEPKKLKGKRDLIVPKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTSGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNRYKSIDGSDETKANWMR 2 1 YVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLAR 1 2 GEKRLQREKSEQVLDNPEDLRGPIHLSVLRQGKSPYKRGFDEGDVHPQAKKKKIDLIFKDVLEASLESAKVEAHQLALSTSLVIRKVPKYQDDAYSQCATTMTHGVQNIGQTQGEGDWKVPQGVSKEPGQLEDEEEEPSSFKADSPAEASLASDPHELPTTSFCPNCIRLKKKVRELQAELDMLKSGKLPEPPVLPPQVLELPEFSDPA 1 2 ASESMVSGPAIMEDDDQEVDSADESVSNDMMTATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSVILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAF SALGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTAATLCEETEFLGDIRAVRWIIGEQN VLNALIKDYLEVVAHLKEVSSQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYIFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAMKNLRVAE AKFQSIREKICQKTQVILAQRFDSRSRIFVKACQVFDLAAWPRSSEELMSYGKEDMVQIFDHLEAIPTFSRDVCREGLDPRGSLLMEWRELKADYYTKNGFKDLISHICKYKQRFPLLNK IIQVLKVLPTSTACCEKGRNALQRVRKNHRSRLTLEQLSDLLTIAVNGPPITNFDAKRALDSWFEEKSGNSYALSAEVLSRMSALEQKPALQTMDHGTEFYPDI* 0 >PRDM4_homSap Homo sapiens (human) 801 aa knuckle, SET, early zinc and array 0 MHHR 2 1 MNEMNLSPVGMEQLTSSSVSNALPVSGSHLGLAASPTHSAIPAP 1 2 GLPVAIPNLGPSLSSLPSALSLMLPMGIGDRGVMCGLPERNYTLPPPPYPHLESSYFRTILP 1 2 GILSYLADRPPPQYIHPNSINVDGNTALSITNNPSALDPYQSNGNVGLEPGIVSIDSRSVNTHGAQSLHPSDGHEVALDTAITMENVSRVTSPISTDGMAEELTMDGVAGEHSQIPNGSRSHEPLSVDSVSN NLAADAVGHGGVIPMHGNGLELPVVMETDHIASRVNGMSDSALSDSIHTVAMSTNSVSVALSTSHNLASLESVSLHEVGLSLEPVAVSSITQEVAMGTGHVDVSSDSLSFVSPSLQMEDSNSNKENMATLFTI 1 2 WCTLCDRAYPSDCPEHGPVTFVPDTPIESRARLSLPKQLVLRQSIVGAEV 1 2 GVWTGETIPVRTCFGPLIGQQSHSMEVAEWTDKAVNHIWK 0 0 IYHNGVLEFCIITTDENECNWMMFVRKAR 2 1 NREEQNLVAYPHDGKIFFCTSQDIPPENELLFYYSRDYAQQI 1 2 GVPEHPDVHLCNCGKECNSYTEFKAHLTSHIHNHLPTQGHSGSHGPSHSKERKWKCSMCPQAFISPSKLHVHFMGHMGMKP HKCDFCSKAFSDPSNLRTHLKIHT 12 GQKN YRCTLCDKSFTQKAHLESHMVIHTGEKN LKCDYCDKLFMRRQDLKQHVLIHTQ 21 ERQ IKCPKCDKLFLRTNHLKKHLNSHEGKRD YVCEKCTKAYLTKYHLTRHLKTCKGPTS SSSAPEEEEEDDSEEEDLADSVGTEDCRINSAVYSADESLSAHK* 0 >PRDM10_homSap Homo sapiens (human) 1160 aa knuckle, SET, early zinc and array 0 MDSKDESSHVWPTSAEHEQNAAQ 0 0 VHFVPDTGTVAQIVYTDDQVRPPQQVVYTADGASYTSVDGPEHTLVYIHPVEAAQ 0 0 TLFTDPGQVAYVQQDATAQQ 0 0 ASLPVHNQVLPSIESVDGSDPLATLQTPLGRLEAKEEEDEDEDEDTEEDEEEDGEDTDLDDWEPDPPRPFDPHDL 1 2 WCEECNNAHASVCPKHGPLHPIPNRPVLTRARASLPLVLYIDRFLGGVFSKRRIPKRTQFGPVEGPLVRGSELKDCYIHLK 0 0 VSLDKGDRKERDLHEDLWFELSDETLCNWMMFVRPAQNHLEQNLVAYQYGHHVYYTTIKNVEPKQELK 0 0 VWYAASYAEFVNQKIHDISEEERK 1 2 VLREQEKNWPCYECNRRFISSEQLQQHLNSHDEKLDVFSR 2 1 TRGRGRGRGKRRFGPGRRPGRPPKFIRLEITSENGEKSDDGTQ 0 0 DLLHFPTKEQFDEAEPATLNGLDQPEQTTIPIPQLPQETQSSLEHEPETHTLHLQPQHEESVVPTQSTLTADDMRRAKRIR 0 0 LELQ 0 0 NAALQHLFIRKSFRP FKCLQCGKAFREKDKLDQHLRFHGREGNCP LTCDLCNKGFISSTSLESHMKLHSDQKT YSCIFCPESFDRLDLLKDHVAIHINDGY FTCPTCKKRFPDFIQ 00 VKKHVRSFHSEKI YQCTECDKAFCRPDKLRLHMLRHSDRKD FLCSTCGKQFK 00 RKDKLREHMQRMHNPEREAKKADRISRSKTFKPRITSTDYDSFT FKCRLCMMGFRRRGML 00 VNHLSKRHPDMKIEEVPELTLPIIKPNRD YFCQYCDK 00 VYKSASKRKAHILKNHPGAELPPSIRKLRPAGPGEPDPMLSTHTQLTGTIATPP VCCPHCSKQYSSK 00 TKMVQHIRKKHPEFAQLSNTIHTPLTTAVISATPAVLTTDSATGETVVTTDLLTQAMTELSQTLTTDYRTPQGDYQRIQYIPVSQSASGLQQPQHIQLQVVQVAS 0 0 ATSPHQSQQSTVDVGQLHDPQPYPQHAIQVQHIQVSEPTASAPSSAQ 0 0 VSGQPLSPSAQQAQQGLSPSHIQGSSSTQGQALQQQQQQQQNSSVQHTYLPSAWNSFRGY 1 2 SSEIQMMTLPPGQFVITDSGVATPVTTGQVKAVTS 0 0 GHYVLSESQSELEEKQTSALSGGVQVEPPAHSDSLDPQTNSQQQTTQYIITTTTNGNGSSEVHITKP* 0 >PRDM15_homSap Homo sapiens (human) 1507 aa knuckle, SET, early zinc finger and intronated array 0 MPRRRPPASGAAQFPERIATRSPDPIPLCTFQRQ 0 0 PRAAPVQPPCRLFFVTFAGCGHRWRSESKPGWISRSRSGIALRAARPP 1 2 GSSPPRPAAPRPPPPGGVVAEAPGDVVIPRPRVQPMRVARGGPWTPNPAFREAESW 2 1 SQIGNQRVSEQLLETSLGNEVSDTEPLSPASAGLRRNPALPP 1 2 GPFAQNFSWGNQENLPPALGKIANGG 1 2 GTGAGKAECGYETESHLLEPHEIPLNVN 0 0 THKFSDCEFPYEFCTVCFSPFKLLGMSGVEGVWNQHSRSASMHTFLNHSATGIREAGCRKDMP 0 0 VSEMAEDGSEEIMFI 12 WCEDCSQYHDSECPELGPVVMVKDSFVLSRAR 2 1 SWPASGHVHTQAGQGMRGYEDRDRADPQQLPEAVPAGLVRRLSGQQLPCRSTLTWGRLCHLVAQGR 2 1 SSLPPNLEIRRLEDGAEGVFAITQLVKRTQFGPFESRRVAKWEKESAFPLK 0 0 VFQKDGHPVCFDTSNEDDCNWMMLVRPAAEAEHQNLTAYQHGSDVYFTTSRDIPPGTELRVWYAAFYAKKMDKPMLKQAGSGVH 1 2 AAGTPENSAPVESEPSQWACKVCSATFLELQLLN 12 EHLLGHLEQAKSLPPGSQSEAAAPEKEQDTPRGEPPAVPESENVATKEQKKKPRRGRKPKVSKAEQPLVIVEDKEPT 12 EQVAEIITEVPPDEPVSATPDERIMELVLGKLATTTTDTSSVPK 21 FTHHQNNTITLKRSLILSSRHGIRRKLIKQLGEHKR VYQCNICSKIFQNSSNLSRHVRSH 12 GDKL FKCEECAKLFSRKESLKQHVSYKHSRNE 00 VDGEYR YRCGTCEKTFRIESALEFHNCRT 12 DDKT FQCEMCFRFFSTNSNLSKHKKKHGDKK FACEVCSKMFYRKDVMLDHQRRHLE 12 GVRRVKblueLEAGGENLVRYKKEP SGCPVCGK 00 VFSCRSNMNKHLLTHGDKK YTCEICGRKFFRVDVLRDHIHVHFK 00 DIALMDDHQREEFIGKIGISSEENDDNSDESADSEPHK YSCKRCQ 00 LTFGRGKEYLKHIMEVHKEKG YGCSICNRRFALKATYHAHMVIHRENLPDPNVQK 21 YI HPCEICGRIFNSIGNLERHKLIHT 12 GVKS HACEQCGKSFARKDMLKEHMRVHDNVRE YLCAECGK 12 GMKTKHALRHHMKLHKGIKE YECKECHRRFAQKVNMLKHCKRHT 12 GIKD FMCELCGKTFSERNTMETHKLIHT 12 VGKQ WTCSVCDKKYVTEYMLQKHVQLTHDKVEA QSCQLCGTKVSTRASMSRHMRRKHPE 0 0 VLAVRIDDLDHLPETTTIDASSIGIVQ 0 0 PELTLEQEDLAEGKHGKAAKRSHKRKQKPEEEAGAPVPEDATFSEYSEKETEFTGSVGDETNSAVQSIQQ 0 0 VVVTLGDPNVTTPSSSVGLTNITVTPITTAAATQFTNLQPVAVGHLTTPERQLQLDNSILTVTFDTVSGSAMLHNRQNDVQIHPQPEASNPQSVAHFINLTTLVNSITPLGSQLSDQHPLTWRAVPQTDVLPPSQPQAPPQQAAQPQVQAEQQQQQMYSY* 0 >PRDM6_homSap Homo sapiens (human) 595 aa knuckle, SET, no early zinc finger, short array 0 MLKPGDPGGSAFLKVDPAYLQHWQQLFPHGGAGPLKGSGAAGLLSAPQPLQPPPPPPPPERAEPPPDSLRPRPASLSSASSTPASSSTSASSASSCAA AAAAAALAGLSALPVSQLPVFAPLAAAAVAAEPLPPKELCLGATSGPGPVKCGGGGGGGGEGRGAPRFRCSAEELDYYLYGQQRMEIIPLNQHTSDPNN 1 2 RCDMCADNRNGECPMHGPLHSLRRLVGTSSAAAAAPPPELPEWLRDLPREVCLCTSTVPGLAYGICAAQRIQQGTWIGPFQGVLLPPEKVQAGAVRNTQHLWE 0 0 IYDQDGTLQHFIDGGEPSKSSWMRYIRCARHCGEQNLTVVQYR 2 1 SNIFYRACIDIPRGTELLVWYNDSYTSFFGIPLQCIAQDEN 1 2 LNVPSTVMEAMCRQDALQPFNKSSKLAPTTQQRSVVFPQTPCSRNFSLLDKSGPIESGFNQINVKNQRVLASPTSTSQLHSEFSDWHL WKCGQCFKTFTQRILLQMHVCTQNPDR 21 P YQCGHCSQSFSQPSELRNHVVTHSSDRP FKCGYCGRAFAGATTLNNHIRTHTGEKP FK 21 CERCERSFTQATQLSRHQRMPNECKP ITESPESIEVD* 0
>ZNF133_homSap Homo sapiens (human) 653 aa KRAB, early zinc finger and array 0 MAFRDVAVDFTQDEWRLLSPAQRTLYREVMLENYSNLVSL 1 2 GISFSKPELITQLEQGKETWREEKKCSPATCP 1 2 DPEPELYLDPFCPPGFSSQKFPMQHVLCNHPPWIFTCLCAEGNIQPGDPGPGDQEKQQQASEGRPWSDQAEGPEGEGAMPLFGRTKKRTLGAFSRPPQRQPVSSRNGLRGVELEASPAQTGNPEETDKLLKRIEVLGFGT VNCGECGLSFSKMTNLLSHQRIHSGEKP YVCGVCEKGFSLKKSLARHQKAHSGEKP IVCRECGRGFNRKSTLIIHERTHSGEKP YMCSECGRGFSQKSNLIIHQRTHSGEKP YVCRECGKGFSQKSAVVRHQRTHLEEKT IVCSDCGLGFSDRSNLISHQRTHSGEKP YACKECGRCFRQRTTLVNHQRTHSKEKP YVCGVCGHSFSQNSTLISHRRTHTGEKP YVCGVCGRGFSLKSHLNRHQNIHSGEKP IVCKDCGRGFSQQSNLIRHQRTHSGEKP MVCGECGRGFSQKSNLVAHQRTHSGERP YVCRECGRGFSHQAGLIRHKRKHSREKP YMCRQCGLGFGNKSALITHKRAHSEEKP CVCRECGQGFLQKSHLTLHQMTHTGEKP YVCKTCGRGFSLKSHLSRHRKTTSVHHR LPVQPDPEPCAGQPSDSLYSL* 0 >HKR1_homSap Homo sapiens (human) 659aa KRAB, early zinc finger and array 0 MRVNHTVSTMLPTCMVHRQTMSCSGAGGITAFVAFRDVAVYFTQEEWRLLSPAQRTLHREVMLETYNHLVSL 1 2 EIPSSKPKLIAQLERGEAPWREERKCPLDLCP 1 2 ESKPEIQLSPSCPLIFSSQQALSQHVWLSHLSQLFSSLWAGNPLHLGKHYPEDQKQQQDPFCFSGKAEWIQEGEDSRLLFGRVSKNGTSKALSSPPEEQQPAQSKEDNTVVDIGSSPERRADLEETDKVLHGLEVSGFGE IKYEEFGPGFIKESNLLSLQKTQTGETP YMYTEWGDSFGSMSVLIKNPRTHSGGKP YVCRECGRGFTWKSNLITHQRTHSGEKP YVCKDCGRGFTWKSNLFTHQRTHSGLKP YVCKECGQSFSLKSNLITHQRAHTGEKP YVCRECGRGFRQHSHLVRHKRTHSGEKP YICRECEQGFSQKSHLIRHLRTHTGEKP YVCTECGRHFSWKSNLKTHQRTHSGVKP YVCLECGQCFSLKSNLNKHQRSHTGEKP FVCTECGRGFTRKSTLSTHQRTHSGEKP FVCAECGRGFNDKSTLISHQRTHSGEKP FMCRECGRRFRQKPNLFRHKRAHSGA FVCRECGQGFCAKLTLIKHQRAHAGGKP HVCRECGQGFSRQSHLIRHQRTHSGEKP YICRKCGRGFSRKSNLIRHQRTHSG* 0 >ZNF343_homSap Homo sapiens (human) 599aa KRAB, early zinc finger and array 0 MMLPYPSALGDQYWEEILLPKNGENVETMKKLTQNHKAK 1 2 GLPSNDTDCPQKKEGKAQIV 0 0 VPVTFRDVTVIFTEAEWKRLSPEQRNLYKEVMLENYRNLLSL 1 2 AEPKPEIYTCSSCLLAFSCQQFLSQHVLQIFLGLCAENHFHPGNSSPGHWKQQGQQYSHVSCWFENAEGQERGGGSKPWSARTEERETSRAFPSPLQRQSASPRKGNMVVETEPSSAQRPNPVQLDKGLKELETLRFGA INCREYEPDHNLESNFITNPRTLLGKKP YICSDCGRSFKDRSTLIRHHRIHSMEKP YVCSECGRGFSQKSNLSRHQRTHSEEKP YLCRECGQSFRSKSILNRHQWTHSEEKP YVCSECGRGFSEKSSFIRHQRTHSGEKP YVCLECGRSFCDKSTLRKHQRIHSGEKP YVCRECGRGFSQNSDLIKHQRTHLDEKP YVCRECGRGFCDKSTLIIHERTHSGEKP YVCGECGRGFSRKSLLLVHQRTHSGEKH YVCRECRRGFSQKSNLIRHQRTHSNEKP YICRECGRGFCDKSTLIVHERTHSGEKP YVCSECGRGFSRKSLLLVHQRTHSGEKH YVCRECGRGFSHKSNLIRHQRTH* 0 >ZNF589_homSap Homo sapiens (human) 364 aa KRAB, early zinc finger and truncated array 0 MWAPREQLLGWTAE 1 2 ALPAKDSAWPWEEKPRYL 0 0 GPVTFEDVAVLFTEAEWKRLSLEQRNLYKEVMLENLRNLVSL 1 2 AESKPEVHTCPSCPLAFGSQQFLSQDELHNHPIPGFHAGNQLHPGNPCPEDQPQSQHPSDKNHRGAEAEDQRVEGGVRPLFWSTNERGALVGFSSLFQRPPISSWGGNRILEIQLSPAQNASSEEVDRISKRAETPGFGAVTFGECALAFNQKSNLFRQKAVTAEKSSDKRQS QVCRECGRGFSRKSQLIIHQRTHTGEKP YVCGECGRGFIVESVLRNHLSTHSGEKP YVCSHCGRGFSCKPYLIRHQRTHTREKS FMCTVCGRGFREKSELIKHQRIHTGDKP YVCRD* 0 >ZNF169_homSap Homo sapiens (human) 603aa KRAB, no early zinc finger but array 0 MSPGLLTTRKEALMAFRDVAVAFTQKEWKLLSSAQRTLYREVMLENYSHLVSL 1 2 GIAFSKPKLIEQLEQGDEPWREENEHLLDLCP 1 2 EPRTEFQPSFPHLVAFSSSQLLRQYALSGHPTQIFPSSSAGGDFQLEAPRCSSEKGESGETEGPDSSLRKRPSRISRTFFSPHQGDPVEWVEGNREGGTDLRLAQRMSLGGSDTMLKGADTSESGAVIRGNYRLGLSKKSSLFSHQKH HVCPECGRGFCQRSDLIKHQRTHTGEKP YLCPECGRRFSQKASLSIHQRKHSGEKP YVCRECGRHFRYTSSLTNHKRIHSGERP FVCQECGRGFRQKIALLLHQRTHLEEKP FVCPECGRGFCQKASLLQHQSSHTGERP FLCLECGRSFRQQSLLLSHQVTHSGEKP YVCAECGHSFRQKVTLIRHQRTHTGEKP YLCPQCGRGFSQKVTLIGHQRTHTGEKP YLCPDCGRGFGQKVTLIRHQRTHTGEKP YLCPKCGRAFGFKSLLTRHQRTHSEEEL YVDRVCGQGLGQKSHLISDQRTHSGEKP CICDECGRGFGFKSALIRHQRTHSGEKP YVCRECGRGFSQKSHLHRHRRTKSGHQL LPQEVF* 0 >ZNF596_homSap Homo sapiens (human) 504 aa KRAB, no early zinc finger but array 0 MPSP 0 0 DSMTFEDIIVDFTQEEWALLDTSQRKLFQDVMLENISHLVSI 1 2 GKQLCKSVVLSQLEQVEKLSTQRISLLQ 1 2 GREVGIKHQEIPFIQHIYQKGTSTISTM 0 0 RSHTQEDPFLCNDLGEDFTQHIALTQNVITYMRTKHFVSKKFGKIFSDWLSFNQHKEIHTKCKSYGSHLFDYAFIQNSALRPHSVTHTREIT LECRVCGKTFSKNSNLRRHEMIHTGEKP HGCHLCGKAFTHCSDLRKHERTHTGEKP YGCHLCGKAFSKSSNLRRHEMIHTREKA QICHLCGKAFTHCSDLRKHERTHLGDKP YGCLLCGKAFSKCSYLRQHERTHNGEKP YECHLCGKAFSHCSHLRQHERSHNGEKP HGCHLCGKAFTESSVLKRHERIHTGEKP YECHVCGKAFTESSDLRRHERTHTGEKP YECHLCGKAFNHSSVLRRHERTHTGEKP YECNICGKAFNRSYNFRLHRRVHTGEKP YVCPLCGKAFSKFFNLRQHERTHTKKAMNM* 0
>GAS8_homSap Homo sapiens (human) synteny marker centromeric to PRDM7 in placentals 0 M 0 0 APKKKGKKGKAKGTPIVDGLAPEDMSKEQ 0 0 VEEHVSRIREELDREREERNYFQLERDKIHTFWEITRRQLEEKKAELRNKDREMEEAEERHQVEIK 0 0 VYKQKVKHLLYEHQNNLTEMKAEGTVVMKLAQKEHRIQESVLRKDMRALKVELKEQELASEVVVKNLRL 0 0 KHTEEITRMRNDFERQVR 1 2 EIEAKYDKKMKMLRDELDLRRKTELHEVEERKNGQIHTLMQRHEEAFTDIKNYYNDITLNNLALINSLK 0 0 EQMEDMRKKEDHLEREMAEVSGQNKRLADPLQKAREEMSEMQKQLANYERDKQILL 0 0 CTKARLKVREKELKDLQWEHEVLEQRFTK 0 0 VQQERDELYRKFTAAIQEVQQKTGFKNLVLERKLQALSAAVEKKEVQFNEVLAASNLDPAALTLVSRKLE 0 0 DVLESKNSTIKDLQYELAQVCK 0 0 AHNDLLRTYEAKLLAFGIPLDNVGFKPLETAVIGQTLGQGPAGLVGTPT >CDH12_homSap Homo sapiens (human) synteny marker chr 5 794 aa MLTRNCLSLLLWVLFDGGLLTPLQPQPQQTLATEPRENVIHLPGQRSHFQRVKRGWVWNQFFVLEEYVGSEPQYVGKLHSDLDKGEGTVKYTLSGDGAGTVFTIDETTGDIHAIRSLDRE EKPFYTLRAQAVDIETRKPLEPESEFIIKVQDINDNEPKFLDGPYVATVPEMSPVGAYVLQVKATDADDPTYGNSARVVYSILQGQPYFSIDPKTGVIRTALPNMDREVKEQYQVLIQAK DMGGQLGGLAGTTIVNITLTDVNDNPPRFPKSIFHLKVPESSPIGSAIGRIRAVDPDFGQNAEIEYNIVPGDGGNLFDIVTDEDTQEGVIKLKKPLDFETKKAYTFKVEASNLHLDHRFH SAGPFKDTATVKISVLDVDEPPVFSKPLYTMEVYEDTPVGTIIGAVTAQDLDVGSSAVRYFIDWKSDGDSYFTIDGNEGTIATNELLDRESTAQYNFSIIASKVSNPLLTSKVNILINVL DVNEFPPEISVPYETAVCENAKPGQIIQIVSAADRDLSPAGQQFSFRLSPEAAIKPNFTVRDFRNNTAGIETRRNGYSRRQQELYFLPVVIEDSSYPVQSSTNTMTIRVCRCDSDGTILS CNVEAIFLPVGLSTGALIAILLCIVILLAIVVLYVALRRQKKKDTLMTSKEDIRDNVIHYDDEGGGEEDTQAFDIGALRNPKVIEENKIRRDIKPDSLCLPRQRPPMEDNTDIRDFIHQR LQENDVDPTAPPYDSLATYAYEGSGSVAESLSSIDSLTTEADQDYDYLTDWGPRFKVLADMFGEEESYNPDKVT* >CDH10 MTIHQFLLLFLFWVCLPHFCSPEIMFRRTPVPQQRILSSRVPRSDGKILHRQKRGWMWNQFFLLEEYTGSDYQYVGKLHSDQDKGDGSLKYILSGDGAGTLFIIDEKTGDIHATRRIDRE EKAFYTLRAQAINRRTLRPVEPESEFVIKIHDINDNEPTFPEEIYTASVPEMSVVGTSVVQVTATDADDPSYGNSARVIYSILQGQPYFSVEPETGIIRTALPNMNRENREQYQVVIQAK DMGGQMGGLSGTTTVNITLTDVNDNPPRFPQNTIHLRVLESSPVGTAIGSVKATDADTGKNAEVEYRIIDGDGTDMFDIVTEKDTQEGIITVKKPLDYESRRLYTLKVEAENTHVDPRFY YLGPFKDTTIVKISIEDVDEPPVFSRSSYLFEVHEDIEVGTIIGTVMARDPDSISSPIRFSLDRHTDLDRIFNIHSGNGSLYTSKPLDRELSQWHNLTVIAAEINNPKETTRVAVFVRIL DVNDNAPQFAVFYDTFVCENARPGQLIQTISAVDKDDPLGGQKFFFSLAAVNPNFTVQDNEDNTARILTRKNGFNRHEISTYLLPVVISDNDYPIQSSTGTLTIRVCACDSQGNMQSCSA EALLLPAGLSTGALIAILLCIIILLVIVVLFAALKRQRKKEPLILSKEDIRDNIVSYNDEGGGEEDTQAFDIGTLRNPAAIEEKKLRRDIIPETLFIPRRTPTAPDNTDVRDFINERLKE HDLDPTAPPYDSLATYAYEGNDSIAESLSSLESGTTEGDQNYDYLREWGPRFNKLAEMYGGGESDKDS >PRDM7_musMus1 Mus musculus genomic strain SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQNSHLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREKp >PRDM7_musMus2 Mus musculus strain WSB/EiJ GU183911 and EU719625 missing a repeat SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQNSHLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREK >PRDM7_musMus3 Mus musculus strain MOLF/EiJ GU183913 SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTEKSSLIQHQRTHTGEKP YVCRECGRGFTQKSNLIKHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP >PRDM7_musMus4 Mus musculus strain PWD/PhJ GU183912 = PWD/Ph FJ212287 SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTEKSSLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP >PRDM7_musMus5 Mus musculus strain CAST/EiJ GU183909 SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSVLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTEKSSLIKHQRTHTGEKP YVCRECGWGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSSLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKPa YVCRECGWGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREK* >PRDM7_musMus6 Mus musculus strain C57BL10.F HQ704390 SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQNSHLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTVKSVLIKHQRTHTGEKP YVCRECGRGFTQNSHLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTQNSHLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREK >PRDM7_musMol Mus musculus molossinus GU216230 SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSVLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTEKSSLIKHQRTHTGEKP YVCRECGWGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSSLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGWGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREK* >PRDM9_musCas Mus musculus castaneus SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSVLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTARSNLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTEKSSLIKHQRTHTGEKP YVCRECGWGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSSLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGWGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREKp >PRDM9_musPah Mus pahari SIERQCGQYFSDKSNVNEHQRTHTGEKP YVCRECGRGFTQKSNLITHQRTHTGEKP YVCRECGRGFTGKSPLIRHQRTHTGEKP YVCRECGRGFTQKSNLITHQRTHTGEKP YVCRECGRGFTGKSPLIRHQRTHTGEKP YVCRECGRGFTQKSHLIKHQRTHTGEKP YVCRECGRGFTEKSNLIKHQRTHTGEKP YVCRECGRGFTQKSPLIRHQRTHTGEKP YVCTECGRGFTQKSNLITHQRTNTGEKP >PRDM9_musMac Mus macedonicus SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTVKSDLIKHQRTHTGEKP YVCRECGRGFTQKSNLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTVKSHLTQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSHLIKHQRTHTGEKP YVCRECGRGFTQKSNLIQHQRTHTGEKP YVCRECGRGFTAKSNLIKHQRTHTGEKP YVCRECGRGFTQNSHLTQHQRTHTGEKS YVCRECGWGFKQKSDLIQHQRTHTREKp >PRDM9_musSpi Mus spicilegus SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQKSNLIQHQRTHTGEK YVCRECGRGFTQKSNLIQHQRTHTGEKP YVCRECGRGFTAKSDLIKHQRTHTGEKP YVCRECGRGFTVKSHLTQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSHLTQHQRTHTGEKP YVCRECGRGFTQKSNLIQHQRTHTGEKP YVCRECGRGFTAKSNLIKHQRTHTGEKP YVCRECGRGFTQNSHLTQHQRTHTGEKS YVCRECGWGFKQKSDLIQHQRTHTREKp >PRDM9_merUng Meriones unguiculatus GTGRECGQCFSDKSNVSEHQRTHTGEKP YVCRECGRGFMQRSNLISHQRTHTGEKP YVCRECGRGFMQRSNLISHQRTHTGEKP YVCRECGRGFTVKSVLISHQRTHTGEKP YVCRECGRGFTVKPHLISHQRTHTGEKP HVCRECGRGFTQRSNLIRHQRTHTGEKP YVCRECGRGFTVKPHLISHQRTHTGEKP YVCRECGRGFTVKPHLISHQRTHTGEKP YVCRECGRGFTVKSVLISHQRTHTGEKP YVCRECGRGFTVKSVLIRHQRTHTGEKP YVCRECRRGFTQRSTLIRHQRTHTGEKP HVCRECGRGFTRGSHLLRHQRTHTGEVL >PRDM9_micAgr Microtus agrestis RVGGERGQCFSDKSNVNEHQRTHTGEKP YVCRECGRGFTRKSNLNVHQRTHTGEKP YVCRECGRGFTRKALLISHQRTHTGEKP YVCRECGRGFTQKALLISHQRTHTGEKP YVCRECGRGFTQKSYLILHQRTHTGEKP YVCRECGRGFTGKSNLNVHQRTHTGEKP YVCRECGRGFTQKSYLILHQRTHTGEKP YVCRECGRGFTGKSLLIRHQRTHTGEKP YVCRECGRGFTQKSYPILHQRTHTGEKp >PRDM9_arvTer Arvicola terrestris RVEGECGQCFNDKSNVNERQRTHTGEKP YVCRECGRGFTRKSVLILHQRTHTGEKP YVCRECGRGFTQKSVLINHQRTHTGEKP YVCRECGRGFTQKSHLIFHQRTHTGEKP YVCRECGRGFTQKSHLILHQRTHTGEKP YVCRECGRGFTWKSVLILHQRTHTGERP YVCRECGRGFTRKSHLILHQRTHTGEKP YVCRECGRGFTQKSHLILHQRTHTGEKP YVCRECGRGFTRKSVLILHQRTHTGEKP YVCRECGRGFTRKSVLINHQRTHTGEKp >PRDM9_perPol Peromyscus polionotus RIETECGQRFSDKSNVNESQRTHSEEKP YVCRECGQGFIQKSVLICHQRTHTGEKP YVCRECGQGFTWKSHLIRHQRTHTGEKP YVCRECGKGFIRKSHLICHQRTHTGEKP YVCRECGQGFIQKSHLICHQRTHTGEKP YVCRECGQGFTQKSVLICHQRTHTGEKP YVCRECGQGFIRKSYLICHQRTHTGEKP YVCRECGKGFTWKSVLIRHQRTHTVEKp >PRDM9_perLeu Peromyscus leucopus RIETECGQRFSDKSNANESQRTHSEEKP YVCRECGQGFTRKSYLICHQRTHTGEKP YVCRECGQGFIQKSVLIRHQRTHTGEKP YVCRECGQGFTRKSYLICHQRTHTGEKP YVCRECGQGFIQKSVLIRHQRTHTGEKP YVCRECGQGFTWKSVLICHQRTHTGEKP YVCRECGQGFTRKSYLICHQRTHTGEKP YVCRECGQGFTWKSHLIRHQRTHTGEKP YVCRECGQGFTRKSYLICHQRTHTGEKP YVCRECGQGFIQKSHLICHQRTHTGEKP YVCRECGQGFTRKSYLICHQRTHTGEKP YVCRECGQGFTWKSVLIRHQRTHTAEKp >PRDM9_perMan Peromyscus maniculatus RTETECGQHFSDKSNANESQRTHSEEKP YVCRECGQGFTWKSVLIRHQRTHTGEKP YVCRECGQGFTWKSVLICHQRTHTGEKP YVCRECGQGFTWKSVLICHQRTHTGEKP YVCRECGQGFIQKSHLIRHQRTHTGEKP YVCRECGQGFIRKSHLICHQRTHTGEKP YVCRECGQGFAQKSVLIYHQRTHTGEKP YVCRECGQGFTRKSHLICHQRTHTGEKP YVCRECGQGFAQKSVLICHQRTHTGEKP YVCRECGQGFTWKSVLICHQRTHTGEKP YVCRECGQGFIQKSHLIRHQRTHTGEKP YVCRECGQGFIQKSHLIRHQRTHTGEKp >PRDM9_apoSyl Apodemus sylvaticus RVERQRGQCFSDKSNVSERQGTHTGEKP CVCRECGRGFTQKSHLNRHQRTHTGEKP HVCRECGRGFTQKSHLNRHQRTHTGEKP HVCRECGRGFTLKSNLNRHQRTHTGEKP CVCRECGRAFTQKSDLIQHQRTHTGEKP YVCRECGRGFTQKSNLNQHQRTHTGEKP YVCRECGRGFTRKSLLIQHQRTHTGEKP YVCRECGRGFTQKSDLNRHQRTHTGEKP YVCRECGRGLTQKSNLIQHQRTHTGEKP YVCRECGRGFTLKSDLIQHQRTHTGEKP YVCRECGRGFTRKSDLNRHQRTHTGEKP YVCRECGRGFTQKSNLIQHQRTHTGEKP YVCRECGRGFTLKSDLIQHQRTHTGEKP YVCRECGRGFTRKSDLNRHQRTHTGEKp >PRDM7_ratNor Rattus norvegicus RIERQCGQCFSDKSNVSEHQRTHTGEKP YICRECGRGFSQKSDLIKHQRTHTEEKP YICRECGRGFTQKSDLIKHQRTHTEEKP YICRECGRGFTQKSDLIKHQRTHTGEKP YICRECGRGFTQKSDLIKHQRTHTEEKP YICRECGRGFTQKSSLIRHQRTHTGEKP YICRECGLGFTQKSNLIRHLRTHTGEKP YICRECGLGFTRKSNLIQHQRTHTGEKP YICRECGQGLTWKSSLIQHQRTHTGEKP YICRECGRGFTWKSSLIQHQRTHTVEKp
Online references
Open 53 recent PubMed abstracts on PRDM9 and related issues. Or use the reverse chronological list below to get free full text for individual articles when that is available:
abs 2012 Sarbajna A major recombination hotspot in the XqYq pseudoautosomal region gives new insight into processing of human gene conversion events. Hum Mol Genet. 2012 Feb 8 htm 2011 Ségurel The Case of the Fickle Fingers: How the PRDM9 Zinc Finger Protein Specifies Meiotic Recombination Hotspots in Humans. PLoS Biol 9(12): e1001211. doi:10.1371/journal.pbio.1001211 htm 2011 Katzman Ongoing GC-Biased Evolution Is Widespread in the Human Genome and Enriched Near Recombination Hot Spots. Genome Biol Evol (2011) 3 614-626 htm 2011 Muñoz Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS One. Nov 2011; 6(11): e25498. htm 2011 Axelsson Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 17 Oct 2011 htm 2011 Grey Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 2011 Oct;9(10):e1001176. 2011 Oct 18. pdf 2011 Berg Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. PNAS 2011 Jul 26;108(30):12378-83. pdf 2011 Richon Chemogenetic analysis of human protein methyltransferases. Chem Biol Drug Des. 2011 Aug;78(2):199-210. abs 2011 Campagna Structural Chemistry of the Histone Methyltransferases Cofactor Binding Site. J Chem Inf Model. 2011 Mar 28;51(3):612-23. pdf 2011 Hinch The landscape of recombination in African Americans. Nature. 2011 Jul 20. doi: 10.1038/nature10336. pmc 2011 Smagulova Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011 Apr 21;472(7343):375-8. htm 2011 Kaupi Distinct properties of the XY pseudoautosomal region crucial for male mouse meiosis. Science 18 Feb 2011;DOI: 10.1126/science.1195774 abs 2011 Briknarova The PR/SET domain in PRDM4 is preceded by a zinc knuckle. Proteins 2011 Jul;79(7):2341-5. doi: 10.1002/prot.23057. pmc 2011 Fledel Variation in human recombination rates and its genetic determinants. PLoS One 2011;6(6):e20321. abs 2011 Neaves Unisexual reproduction among vertebrates. Trends Genet. 2011 Mar;27(3):81-8. abs 2011 Ponting What are the genomic drivers of the rapid evolution of PRDM9? Trends Genetics (2011) 1–7 htm 2011 Yanover Extensive protein and DNA backbone sampling improves structure-based specificity prediction for C2H2 zinc fingers. Nucleic Acids Res. 2011 Feb 22 pdf 2011 Ubeda Red Queen theory of recombination hotspots. J Evol Biol. 2011 Mar;24(3):541-53. abs 2010 Hochwagen Meiosis: a PRDM9 guide to the hotspots of recombination. Curr Biol. 2010 Mar 23;20(6):R271-4. pmc 2010 Tsai Conservation of recombination hotspots in yeast. PNAS 2010 April 27; 107(17): 7847–7852. abs 2010 Klug The discovery of zinc fingers and practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010 Feb;43(1):1-21. abs 2010 Berg PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010 Oct;42(10):859-63. abs 2010 McVean PRDM9 marks the spot. Nat Genet. 2010 Oct;42(10):821-2. pdf 2010 Kong Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010 Oct 28;467(7319):1099-103. pmc 2010 Parvanov Prdm9 controls activation of mammalian recombination hotspots. Science. 2010 Feb 12;327(5967):835. pmc 2010 Lorenz The ancient mammalian KRAB zinc finger gene cluster on human chromosome 8q24.3 BMC Genomics. 2010 Mar 26;11:206. pmc 2010 Neale PRDM9 points the zinc finger at meiotic recombination hotspots. Genome Biol. 2010;11(2):104. pmc 2010 Sandovici PRDM9 sticks its zinc fingers into recombination hotspots and between species. F1000 Biol Rep. 2010 May 24;2. pmc 2010 Billings Patterns of recombination activity on mouse chromosome 11 revealed by high resolution mapping. PLoS One. 2010 Dec 8;5(12):e15340. htm 2010 Cheung Genetic control of hotspots. Science. 2010 Feb 12;327(5967):791-2. pdf 2010 Urnov Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005 Jun 2;435(7042):646-51. htm 2010 Zheng Detecting sequence polymorphisms associated with meiotic recombination hotspots in the human genome. Genome Biol. 2010;11(10):R103. htm 2010 Baudat PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010 Feb 12;327(5967):836-40. htm 2010 Myers Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010 Feb 12;327(5967):876-9. pmc 2009 Jeffreys The rise and fall of a human recombination hot spot. Nat Genet 2009 41:625–629. pmc 2009 Berglund Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009 Jan 27;7(1):e26. pmc 2009 Thomas Evolution of C2H2-zinc finger genes revisited. BMC Evol Biol. 2009 Mar 4;9:51. pmc 2009 Oliver Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009 Dec;5(12):e1000753. pmc 2009 Thomas Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS One. 2009 Dec 30;4(12):e8505. htm 2009 Willis Origin of species in overdrive. Science. 2009 Jan 16;323(5912):350-1. htm 2009 Irie Single-nucleotide polymorphisms of the PRDM9 (MEISETZ) gene in patients with nonobstructive azoospermia. J Androl. 2009 Jul-Aug;30(4):426-31. htm 2009 Mihola A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009 Jan 16;323(5912):373-5. abs 2008 Brayer The protein-binding potential of C2H2 zinc finger domains. Cell Biochem Biophys. 2008;51(1):9-19. htm 2008 Webb Sperm cross-over activity in regions of the human genome showing extreme breakdown of marker association. PNAS 2008 105:10471–10476 pmc 2008 Duret The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008 May 9;4(5):e1000071. pmc 2008 Miyamoto Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet. 2008 Nov-Dec;25(11-12):553-7. htm 2008 Cho Prediction of DNA binding sites for zinc finger proteins. BBRC 2008 May 9;369(3):845-8. abs 2008 Myers A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008 Sep;40(9):1124-9. pmc 2007 Coop Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007 Mar 9;3(3):e35. pmc 2007 Fumasoni Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007 Oct 4;7:187. htm 2007 Gay Estimating meiotic gene conversion rates from population genetic data. Genetics. 2007 Oct;177(2):881-94. pdf 2006 Phillips A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis. Dev Cell. 2006 Dec;11(6):817-29. htm 2006 Birtle Meisetz and the birth of the KRAB motif. Bioinformatics. 2006 Dec 1;22(23):2841-5. pdf 2006 Hayashi Meisetz, a novel histone tri-methyltransferase, regulates meiosis-specific epigenesis. Cell Cycle. 2006 Mar;5(6):615-20. pdf 2005 Hayashi A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005 Nov 17;438(7066):374-8. htm 2005 Winckler Comparison of Fine-Scale Recombination Rates in Humans and Chimpanzees. Science 2005 2005 Apr 1;308(5718):107-11. abs 2000 Laity DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J Mol Biol. 2000 Jan 28;295(4):719-27.
Article author
I researched this article in its entirety in April and July-August of 2011, paying as little attention as possible to the previous studies above, which are excellent on meiosis but completely clueless on comparative genomics. This is a moderately difficult topic as human genes go, so the overall annotation is still being revised periodically into 2012 as better quality genomes become available. A change in one section places others in need of revision, not always promptly attended to.
Although copyrighted, all the information here is in the public domain and can be used by anyone without additional permissions if properly sourced; however if data, figures or original observations are taken for a peer-reviewed scientific publication, it might be appropriate (after consultation early on) to include me among secondary co-authors.
Rather than make article edits yourself, please contact me by email with clarifications, corrections or additions to the content so I can make edits while maintaining a consistent approach. For broader disagreements or different interests, a better option is to register at the UCSC genomeWiki site and create your own page within the comparative genomics category.
This is just a scientific research article on a vertebrate gene family, not a counseling resource for personal genomics nor medical advice on infertility -- thanks in advance for not sending inappropriate email. Note technical terms from genetics and molecular biology are not explained when keywords have a satisfactory treatment at wikipedia or in undergraduate genetics texts.
My last dozen published research papers in PNAS, Nature, Science etc can be found here. Watch for 4 additional comparative genomics paper to appear in 2012. I've also written over a thousand pages of comparative genomics for other human genes, authored the original user manual to the UCSC human genome browser and in 1999 an advanced tutorial on metazoan genome annotation still widely available online. I thank the UCSC Genomics Group (Hiram Clawson, Brian Raney) for software and manuscript resources, Evim Foundation for logistical support, and the Sperling Foundation for financial support under project grant 2011.GNTCS.004.