Gene Set Summary Statistics: Difference between revisions
From genomewiki
Jump to navigationJump to search
| Line 106: | Line 106: | ||
txEnd-txStart)</TH></TR> | txEnd-txStart)</TH></TR> | ||
<TR><TH>hg17</TH> | <TR><TH>hg17</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr4: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr4:69806230-69806397 AF241539] (168)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7:129369319-129369494 AF277175] (176)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX:9353026-9353265 AY459291] (240)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr1: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr1:35869683-35869925 AY605064] (243)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7:128643905-128644162 AF503918] (258)</TD> | ||
</TR> | </TR> | ||
<TR><TH>hg18</TH> | <TR><TH>hg18</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr9: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr9:130062323-130062342 uc004buj.1] (20)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr1: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr1:65874524-65874545 uc001dcm.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr12: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr12:52671813-52671834 uc001seo.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr12: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr12:56504708-56504729 uc001sqn.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr20: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr20:15523127-15523148 uc002wpa.1] (22)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm8</TH> | <TR><TH>mm8</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chrX:12185020-12185236 AJ319753] (217)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr3: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr3:92585666-92585896 BC107019] (231)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr11: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr11:70546334-70546619 BC016221] (286)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr16: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr16:88757957-88758259 NM_130876] (303)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr16: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr16:88773645-88773948 NM_130873] (304)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm9</TH> | <TR><TH>mm9</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr1: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr1:74440898-74440919 uc007bma.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr10: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr10:85230424-85230445 uc007gmr.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr11: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr11:77886549-77886570 uc007khz.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr12: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr12:110831098-110831119 uc007pay.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr13: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr13:54944553-54944574 uc007qpn.1] (22)</TD> | ||
</TR> | </TR> | ||
</TABLE> | </TABLE> | ||
==Custom Track of Small Exons and Introns== | ==Custom Track of Small Exons and Introns== | ||
Revision as of 17:28, 19 September 2007
gene sets measured
- hg17 - knownGenes version 2
- hg18 - knownGenes version 3
- mm8 - knownGenes version 2
- mm9 - knownGenes version 3
The min, max and mean measurements are per gene
summary of gene and exon counts
| db | gene count | total exon count | min exon count | max exon count | mean exon count |
|---|---|---|---|---|---|
| hg17 | 39368 | 405720 | 1 | 149 | 10 |
| hg18 | 56722 | 519308 | 1 | 2899 | 9 |
| mm8 | 31863 | 314628 | 1 | 313 | 10 |
| mm9 | 49220 | 417114 | 1 | 610 | 8 |
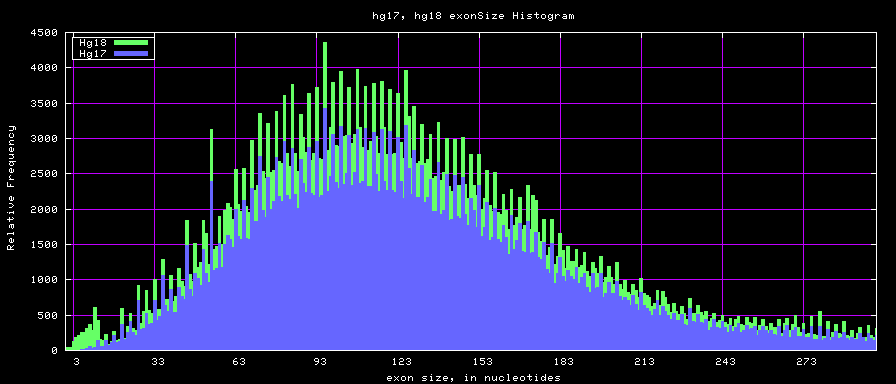
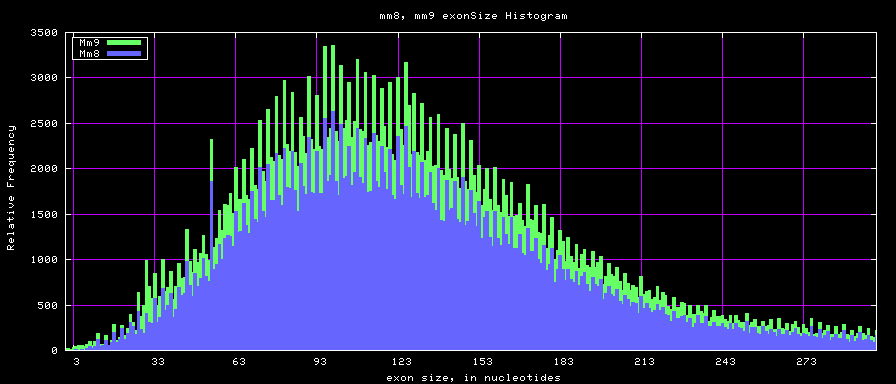
summary of exon size statistics
| db | sum exon sizes | min exon size | max exon size | mean exon size |
|---|---|---|---|---|
| hg17 | 106839720 | 1 | 18172 | 263 |
| hg18 | 146371091 | 1 | 36861 | 282 |
| mm8 | 83159087 | 4 | 17497 | 264 |
| mm9 | 117671086 | 1 | 29698 | 282 |
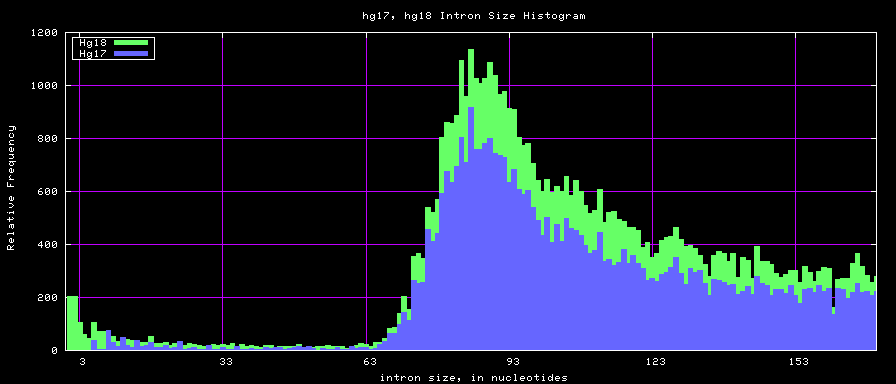
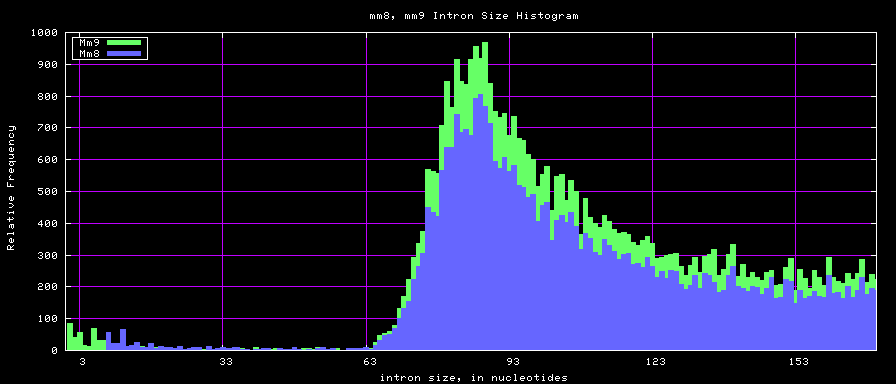
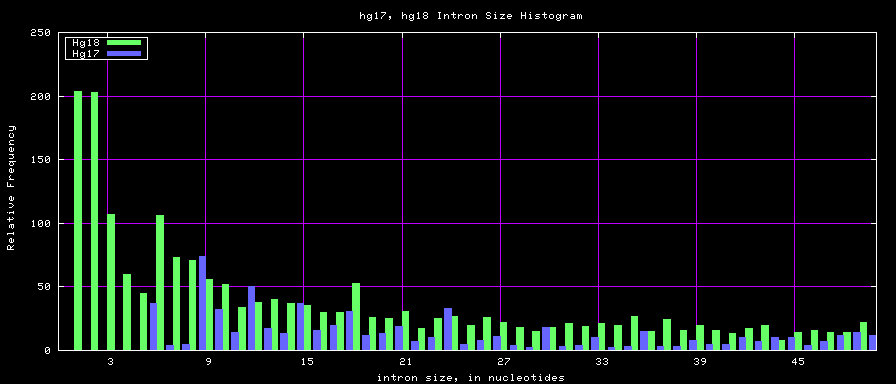
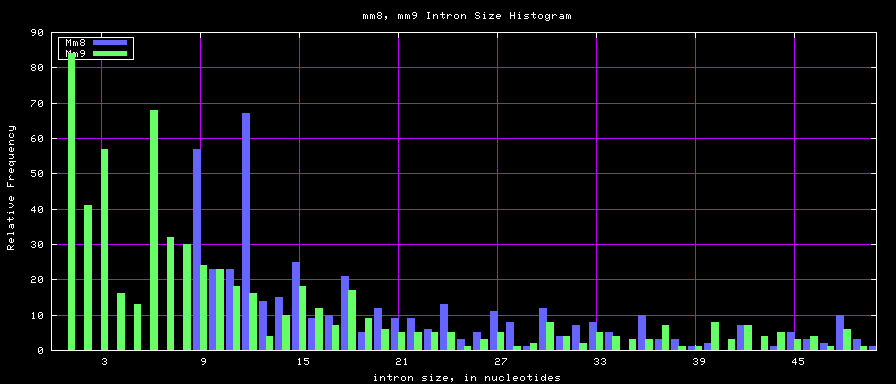
summary of intron size statistics
| db | sum intron sizes | min intron size | max intron size | mean intron size |
|---|---|---|---|---|
| hg17 | 2223224397 | 6 | 1096450 | 6069 |
| hg18 | 2784923600 | 1 | 1047320 | 6023 |
| mm8 | 1476081990 | 9 | 1347550 | 5220 |
| mm9 | 2055504784 | 1 | 1253430 | 5589 |
Top five exon count genes
| db | gene name (exon count) | ||||
|---|---|---|---|---|---|
| hg17 | NM_004543 (149) | AF535142 (146) | AF535142 (146) | NM_033071 (146) | AF495910 (146) |
| hg18 | uc001yrq.1 (2899) | uc002zvw.1 (322) | uc002umr.1 (313) | uc002stk.1 (217) | uc002umt.1 (194) |
| mm8 | NM_011652 (313) | NM_028004 (192) | NM_007738 (118) | NM_134448 (99) | DQ067088 (99) |
| mm9 | uc007pgj.1 (610) | uc008kfn.1 (313) | uc008kfo.1 (192) | uc008jqv.1 (157) | uc009rrh.1 (118) |
Top five largest CDS extent genes
| db | gene name (CDS extent size: thickEnd-thickStart) | ||||
|---|---|---|---|---|---|
| hg17 | NM_014141 (2298740) | NM_000109 (2217347) | CR749820 (2138880) | NM_004006 (2089394) | X14298 (2089394) |
| hg18 | uc003weu.1 (2298740) | uc004ddb.1 (2217347) | uc001pak.1 (2138880) | uc004dda.1 (2089394) | uc003wqd.1 (2055833) |
| mm8 | NM_007868 (2253366) | NM_001004357 (2238304) | NM_053011 (2055883) | AK134694 (1988713) | NM_053171 (1639258) |
| mm9 | uc009tri.1 (2253366) | uc009bst.1 (2238325) | uc007zfr.1 (2189582) | uc008jon.1 (2055883) | uc008mpv.1 (1988713) |
Top five smallest transcript genes
| db | gene name (transcript size: txEnd-txStart) | ||||
|---|---|---|---|---|---|
| hg17 | AF241539 (168) | AF277175 (176) | AY459291 (240) | AY605064 (243) | AF503918 (258) |
| hg18 | uc004buj.1 (20) | uc001dcm.1 (22) | uc001seo.1 (22) | uc001sqn.1 (22) | uc002wpa.1 (22) |
| mm8 | AJ319753 (217) | BC107019 (231) | BC016221 (286) | NM_130876 (303) | NM_130873 (304) |
| mm9 | uc007bma.1 (22) | uc007gmr.1 (22) | uc007khz.1 (22) | uc007pay.1 (22) | uc007qpn.1 (22) |
Custom Track of Small Exons and Introns
Custom track: Hg18 small exons and introns on the UCSC Genes track
These are exons of size less than 22 bases, and introns of size less than 12 bases. The score column contains the size and thus you can filter smaller subsets via the score column in the table browser.
These small exons and introns are used to maintain frame coding boundaries as found in mRNAs compared to the reference genome coordinates.
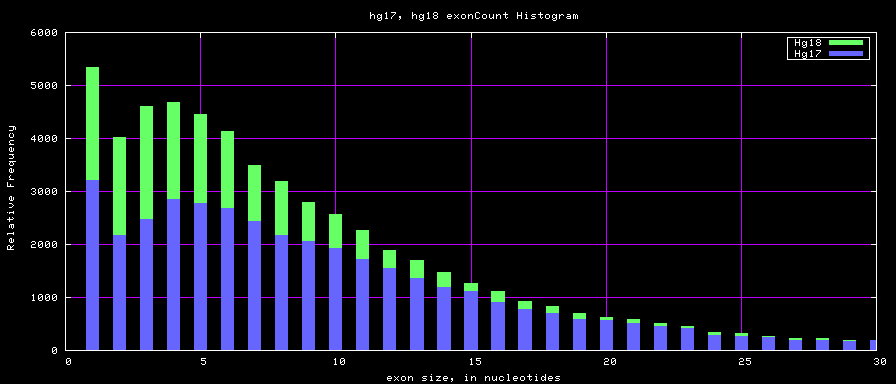
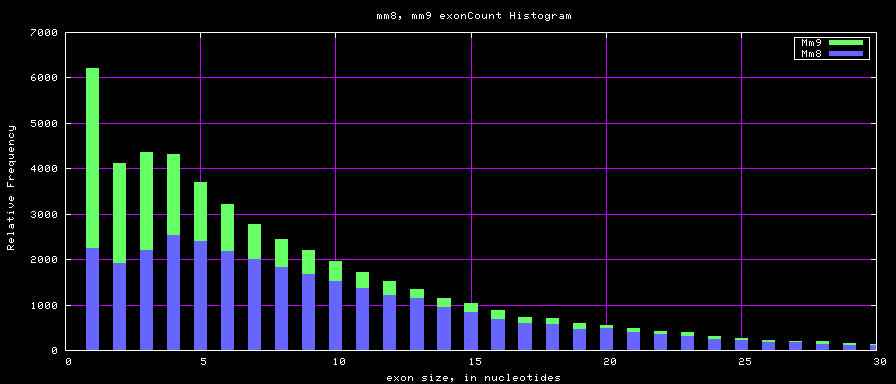
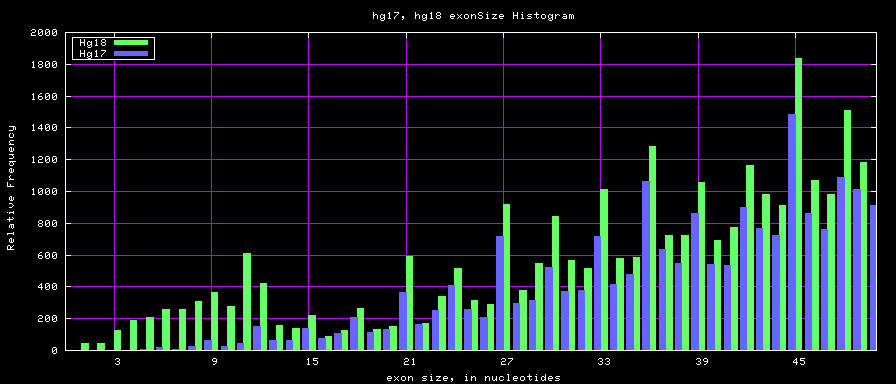
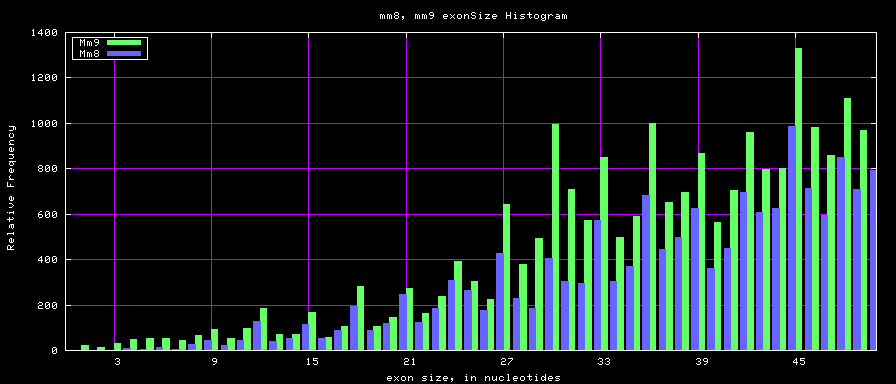
Histogram graphs
Methods
- From the table browser, request three different bed files for the knownGenes track:
- whole gene
- exons only
- introns only
- From those bed files, stats can be extracted
- gene count from: 'wc -l wholeGene.bed'
- exon count stats from:
STATS=`ave -col=10 wholeGene.bed -tableOut | grep -v "^#"`
MIN=`echo $STATS | cut -d' ' -f1`
MAX=`echo $STATS | cut -d' ' -f5`
MEAN=`echo $STATS | cut -d' ' -f6 | awk '{printf "%d", $1+0.5}'`
COUNT=`echo $STATS | cut -d' ' -f8 | awk '{printf "%d", $1}'`
- for exon or intron size stats:
STATS=`awk '{print $3-$2}' {introns,exons}.bed \
| ave -col=1 stdin -tableOut | grep -v "^#"`
MIN=`echo $STATS | cut -d' ' -f1`
MAX=`echo $STATS | cut -d' ' -f5 | awk '{printf "%d", $1}'`
MEAN=`echo $STATS | cut -d' ' -f6 | awk '{printf "%d", $1+0.5}'`
SUM_SIZE=`awk '{sum += $3-$2} END{printf "%d", sum}' {introns,exons}.bed`
- top five exon count genes
sort -k10nr wholeGene.bed | head -5
- top five CDS size genes
awk '{cdsSize=$8-$7
if (cdsSize > 0) {printf "%s\t%s\t%s\t%s\t%d\n", $1,$2,$3,$4,cdsSize}
}' wholeGene.bed | sort -k5nr | head -5
- top five smallest transcript genes
awk '{size=$3-$2
if (size > 0) {printf "%s\t%s\t%s\t%s\t%d\n", $1,$2,$3,$4,size}
}' wholeGene.bed | sort -k5n | head -5