Opsin evolution: Difference between revisions
Tomemerald (talk | contribs) No edit summary |
Tomemerald (talk | contribs) No edit summary |
||
| Line 1,289: | Line 1,289: | ||
1 PHSEMFIHPLLAEIPVMMAKASARYNPIIYALSHPKFR 2 | 1 PHSEMFIHPLLAEIPVMMAKASARYNPIIYALSHPKFR 2 | ||
1 AEIDKHFPWLLCCCKPKPKAQLPSSTTKGSIASKTEADTSV* 0 | 1 AEIDKHFPWLLCCCKPKPKAQLPSSTTKGSIASKTEADTSV* 0 | ||
>MEL_helRob Helobdella robusta (leech) fragmentary model from scaffold_39 | |||
1 TPILRTHANVLIINLALCDLIFSSLIGFPMTALSCFKRHWIWGDL 1 | |||
2 GCDFYGFVAGWTGLGSITCLAFISIDRYMAIVHPFYMLNKKSSSVLTLLGCDFYGFVAGWTGLGSITCLAFISIDRYMAIVHPFYMLNKKSSSVLTLLIIVTSYIGIVIEVTKS 1 | |||
1 KELKTAKVLACCFGAFLICWTPYAIVAQLGINGFAHLVTPFTSEVPVLFAKTSSIWNPLIYALSHPRYRRAV 0 | |||
>MOLL_RHO_lolSub Loligo subulata Z49108 499 Mollusca Cephalopoda complete | >MOLL_RHO_lolSub Loligo subulata Z49108 499 Mollusca Cephalopoda complete | ||
Revision as of 18:33, 5 December 2007
Below is a large set of phylogenetically representative hand-curated intronated opsin sequences that serves as a gene family classifier ... just uBlast an unknown sequence against the database below and look for consistent labelling of the top hits from the Opsin Classifier. It takes only 6 seconds per query!
The set of sequences is not intended to be exhaustive. Rather, if a given clade has many available similar sequences, those with genome assemblies are chosen to represent the group, for example anole is preferred to gecko, and (rightly or wrongly) any experimental results transfered over. This avoids uninformative clutter from near-identical sequences. If the clade reflects a very deep divergence such as lamprey or amphioxus, all available sequences are provided so as to break up long branches.
About half the sequences are not available from GenBank but rather are culled from trace archives (see tutorial), genomic contigs, and genome assemblies, typically by blastx against the full (and growing) set of reference sequences. The level of error in the curated sequences is very low, declines with time as anomalies are revisited and fixed, but never reaches zero because of problems inherent to experimental data, incomplete assemblies, and sequence manipulation.
Thus the usual querying at GenBank does not remotely approach the basic capabilities of the Opsin Classifier. Those sequences are spread out over many separate databases, not accessible by any single method, often misannotated and/or mislabelled by some unattended pipeline , with edge creep of genomic matches difficult to read, uncorrected frameshifts, unnecessary truncation, and erroneous amino terminals.
As worst case example, half-baked annotation of the sea urchin genome by pipelines and casual procedures has left a terrible legacy of erroneous opsin gene structures at GenBank, journals, and genome browsers -- often mis-classified as well because of an inadequate set of reference sequences, chimeric confusion in tandem duplicates, and non-consideration of intron structure and synteny. These errors could trigger additional downstream errors for anyone using GenBank nr as classifier. This may eventually lead to wholly non-existent "virtual opsins" with attending vacuous but seemingly documented speculation on echinoderm photoreception roles.
The fasta header of each sequence is a miniature database, with fields showing the opsin type, genus, species and common name, accession number, best PubMed citation, indels, intron pattern, sequence length, lambda max adsorption, flanking synteny, and G protein type with which it interacts (all subject to availability and work-in-progress). These novel fasta headers by themselves provide a quick over view the collection -- simply paste into a blank document and pull lines containing '>'.
The protein sequences are broken into their constituent exons using genomic information when available. When not available (eg the opsin originated as a cDNA in a species lacking a genome project), the exons are inferred from the phylogenetically closest opsins. The numbers flanking exons, 012, show the phasing of each intron, eg 12 means an overhang of 1 bp at the 3' end of an exon with that fragmentary codon completed by a 2 bp overhang at the beginning of the next exon. Intron position and phasing are generally conserved over great evolutionary distances -- note here lamprey eel has identical intronation of its opsin genes orthologous to human. Cone and rod opsin paralogs are intronated identically in all species with the exception of LWS opsins which have an extra early intron of phase 12. LWS must have acquired this prior to divergence of lamprey.
Syntentic relationships are also shown. The nearest flanking HUGO-named genes are first chosen for the human opsin, two on each side. The strand orientation noted relative to a fixed convention of plus strand for the human opsin. Then each assembly is revisited to determine the extent of conservation of these flanking genes. In the event humans lack the gene, synteny is defined by the nearest diverging species, typically platypus, that has the gene. Sometimes the original synteny is only partly retained (left- or right-synteny). For deeply diverging species such as amphioxus with an assembly, flanking genes there are pushed forward into other species to help define orthologous opsins (blast clustering can be uncertain because of the diminishing percent identity).
Melanopsins, the unexpected rhabdomeric-class Gq-coupled opsin recently found in upper deuterostomes, are readily confused homologically due to various expansions and contractions. Mammals, human through platypus, have a single melanopsin. However chicken, lizard, frog, and teleost fish experienced a multi-gene segmental duplication and the resulting melanopsins were both retained (though diverged substantially). In ray-finned fish, a processed retrogene arose that may be functional in zebrafish though lost in fugu and stickleback. After its whole genome duplication, zebrafish also retained two copies of the original melanopsin. Chondrichthyes also have a second copy of the primary melanopsin but synteny -- which is essential for analysis since intron placement is uninformative in duplications and sequence alignment is too dependent on unknown rates -- is not available in the current contig-level assembly.
Amphioxus also contains two melanopsins from an apparently independent duplication. Flanking gene order today bears no relation to vertebrate gene order. The lamprey situation awaits assembly of its traces or targeted transcript studies. At this time, only a four exon fragmentary melanopsin can be recovered (however with high percent identity, 80%). Possibly orthologs of this melanopsin locus could be tracked into the highly derived tunicates, acorn worm, and sea urchins. The distinctive intron pattern may even allow melanopsin antecedents to be identified in Cnidaria and Protostomia. At this point, the best blastp match to insects stands at 37% with no evident syntenic or intronic support
While clade-specific proliferation of melanopsins -- and implied role subfunctionalization -- confounds the situation for chordates, it really has little impact on the opsin classifier described here. Unknown sequences will readily find their place because of excellent phylogenetic distribution of reference sequences and the inherent distance of melanopsins from the ciliary collection. The main utility at the level of opsin classifier is the ability to identify other rhabdomeric opsins in later deuterostomes should they occur. At the level of alignment, the melanopsis serve as outgroup to ciliary opsins and so help define motifs specific to Gt-coupled signaling and other structure/function issues.
A dozen very recent publications have shaken our understanding of the evolution of light reception capabilities. After reviewing topics such as ciliary opsin in protostomes, rhabdomeric opsins in deuterostomes, rich opsin repertoires in cnidarians, and other novel opsin classes, I will consider topics such as the origin of image-forming eyes beween amphioxus and lamprey divergences, noting however that our notion of 'eye' is much more nuanced today. The reconstruction of the ur-bilateran eye probably awaits additional cnidarian genomes -- no new ones are being undertaken unfortunately. However the plethora of new arthopod and lophotrochozoan genome assemblies has opened up new avenues of research as the realization grows that fly and nematode are exceedingly derived, with better ancestral characters retained in other species.
Numerous conflicting gene trees have been published for ciliary opsins. Some methodologies have bordered on the preposterous -- thin phylogenetic coverage, dimly related outgroups such as drosophila rhabdomeric opsin, and naive fixed underlying mutational models assumed for maximal likelihood software despite the great diversity of species and many billions of years of branch length. Nonetheless, the resultant trees have only moderate conflict, suggesting that a definitive opsin tree might not be far off.
Rare genomic changes have lately come into vogue as a supplement to traditional maximal likelihood methods, primarily to resolve polytomies (divergence nodes tightly spaced close in time) and otherwise uncertain gene or species tree topologies. The rare genomic changes applicable to opsins include coding indels (deletions and insertions), intron placement (position and phase comparison), synteny (gene order along the chromosome), and gene copy number change (gene gain from retropositional, tandem, segmental, and whole genome duplications; gene loss from pseudogenization or deletion). Results from these methods must be evaluated for their susceptibility to homoplasy (misleading recurrent independent events that mimic a single event) and incomplete penetration in the population level at the time of speciation (lineage sorting).
Among other phylogenetically informative rare genomic events, we'll be looking at a 6 bp amino acid insert, a novel 12 upstream intron in LWS, and post-GWSR introns in rod/cone opsins, all events located between transmembrane helices TM2 and TM3, ie in extracellular loop 2. Their lack of homoplasy can be seen in the massive alignments below.
Because not all cDNA sequences takes place in species having genome projects and not all species having genome projects have cDNAs, existing cDNAs had to be aligned within the heterologous genome project in order to determine their intron placement. As an example, lamprey opsins from Geotria australis and Lethenteron japonicum worked as queries to locate orthologs within the Petromyzon maritimus genome project (which consists solely of 19 million traces as of mid-November 2007).
The first point to be understood in ciliary opsin evolution is jawless fish such as lamprey exhibit a full-blown set of modern rod and cone opsins whereas early deuterostomes such as hemichordates, echinoderms, amphioxus and tunicates genomes totally lack them (Xenoturbella is not available yet) and indeed altogether lack conventional imaging eyes while using protostome-like rhabdomeric opsins with their disjunct signaling system for photorecepton. Of course, characters in extant (living) species should never be confused with ancestral characters at the time of divergence nodes (last common ancestors); conceivably these early diverging deuterostomes have lost opsin genes, perhaps due to a habitat shift to deep water or burrowing habitat.
However the molecular evidence is quite clear that full-blown pentachromatic color vision and most other modern ciliary opsin classes first appeared during the evolutionary stem preceding lamprey divergence. The oldest known fossil lamprey, Priscomyzon, dates at 360 myr to the Devonian. Molecular clocks place lamprey appearance at approximately 430 myr, some 100 million years after Chengjiang and Burgess Shales fossil Lagerstatte formed. Like most soft tissues, eyes seldom leave a good fossil record, though bilateral placement might be reflected in bone orbits.
Hagfish, sister group to lamprey, have imaging eyes but have not been studied; their opsins situation may be derived due to deepwater marine habitat (similarly deepwater coelocanth opsins are adapted to 420 nm). The next-diverging chondrichtyes have inadequate data at GenBank -- only a few rhodopsin genes from skates and dogfish.
This makes even fragments from the partially sequenced elephantfish Callorhyncus milii quite valuable. Those 9 fragments and 3 from the lamprey genome are provided in the data section. The opsin classifier tool can reliably type a fragment from a single mid-sized exon. While full length genes are always preferable, these fragments serve to prove existence of that gene class at the time of a given divergence node. Further, they can validate certain rare genomic events provided the fragment happens to overlap the region of interest.
Despite 6 sequenced opsin mRNAs in the amphioxus Branchiostoma belcheri and an initial assembly in Branchiostoma floridae, no rod/cone opsin can be located there or in earlier diverging deuterostomes with genome projects (3 unicates, 2 urchins, 1 acorn worm). These species may have larval eye spots, ocelli, pigment cells, and related photoreceptors but lack imaging eyes.
The fossil record is unsatisfactory: less than 1 bilateran in 10,000 in Chengjiang and Burgess Shale fossils is even a candidate for deuterostomy. Low numbers of specimens and poor preservation conspire with career pressure and cite-seeking journals to egregiously misinterpret data in the analysis of Hou, discoverer of the Chinese lagerstaette. Myllokunmingia is in the best situation with 500 specimens but Haikouichthys as stem deuterostome, Metaspreggina as post-ediacaran, and Yunnanozoan are all problematic (in the eye of the beholder). While signs of bilaterily disposed eyes are sometimes inferred, it does not follow these were image-forming eyes. Indeed contemporary Branchiostoma and tunicate larva have an eye-spot (ocellus); the genomes contain ciliary opsins clustering to approximately ENCEPH and PPIN -- still a long long road to imaging opsins. Echinoderms and hemichordates genomes have opsins but even more remote. Sea urchin genome encodes at least six opsins, four of these cluster classify to rhabdomeric, ciliary and Go-type. Tube feet are apparently the photosensory organ in adult urchins.
Meanwhile, thousands of high-quality Cambrian arthropod fossils unmistakably show stalked paired eyes. Fossil trilobite eyes are much studied, due to use of calcite as lens crystalin. Imaging eyes of contemporary arthropods and lophotrochozoa are rhabdomeric, utilizing depolarizing Gq-type receptor, phospholipase C, phosphoinositola, diacylglycerol, and transient receptor potential TRP and TRPL channel signaling. However their genomes can also contain ciliary opsins, using hyperpolarizing Gt-type transducins and phosphodiesterase cGMP second-messaging (as well as Go-type gustducin ciliary opsins in other types of photoreceptors).
Vertebrates are just the opposite, having crossed over to a ciliary opsin-based imaging system, while retaining rhabdomeric signaling in melanopsin retinal ganglion cells. Cnidarian opsins are available from Hydra and Nematostella genomes. Hydra expresses a ciliary-type opsin in ectodermal sensory nerve cells whereas Nematostella has opsins classifying between melanopsin and encephalopsin.
It must not be thought that bilaterans invented imaging eyes because earlier diverging cubomedusan jellyfish Carybdea marsupialis has 4 eyestalks each with 6 photoreceptors of 4 types: simple eyespots, pigment cups, complex pigment cups with lenses, and camera-type eyes with a cornea, lens, and retina. This jellyfish tracks, captures, and eats teleost fish. The species very much needs a genome project.
Thus there is no evidence whatsoever -- and every reason to doubt from genomic analysis -- that deuterostomes had imaging eyes during the Cambrian. Despite this, a BBC series, Walking With Monsters, portrayed a school of 25 mm Haikouichthys attacking and wounding an Anomalocaris twenty times their size. It is easy to guess at the scientific advisory panel. This recurrent anthropocentric theme is echoed by fantastic museum imagery of early mammals nimbly predating on dinosaur nests -- dioramas quietly dismantled after Yucatan meteriorite discovery.
Imaging eyes are not essential to survival; even today subterranean mammals such as blind mole rat flourish without them. Discounting ray-finned fish numbers, a very substantial proportion of extant animal species lack imaging eyes 525 myr after the Cambrian. Of 33 animal phyla, a one-third have no specialized organ for detecting light, one-third have light-sensitive organs, and the remaining 6 have imaging eyes (Cnidaria, Mollusca, Annelida, Onychophora, Arthropoda, and Chordata). Thus 82% of animal phyla have survived well over 500 myr without imaging eyes despite the supposedly unrelenting competition/predation from animals with them.
The first table below shows the reference opsin sequences at a glance, grouped by class. Below that is the primary collection of opsing protein sequences. Here the "fields" in the fasta header show gene name, genus, species, common name, heterotrimeric G protein alpha subunit used in signaling, intron structure, synteny (2 flanking genes on each side of the opsin), indel status, sequence length, lambda max, and comment field.
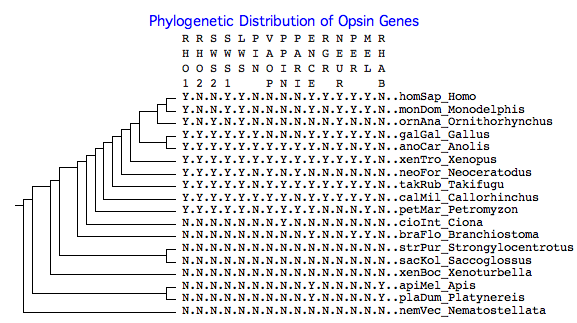
The phylogenetic tree below shows the presence or absence of various opsin genes in clade-representative species, as reflected in the collected reference sequences. The purpose is timing appearance (or disappearance) of a given class of opsin gene. For example, cone and rod opsins first appeared before lamprey divergence; otherwise they are absent from urochordates, cephalochordates, and earlier deuterostomes. Note however a given gene might appear absent because of a genome project gap, lack of experimental effort, insufficient or outdated bioinformatics, or species idiosyncracies (ie be present in a different species of that clade). In other cases (eg platypus SWS1) pseudogene remnants or a syntenically proven deletion establish the gene is definitely absent. Y means yes (present), N means no (absent).
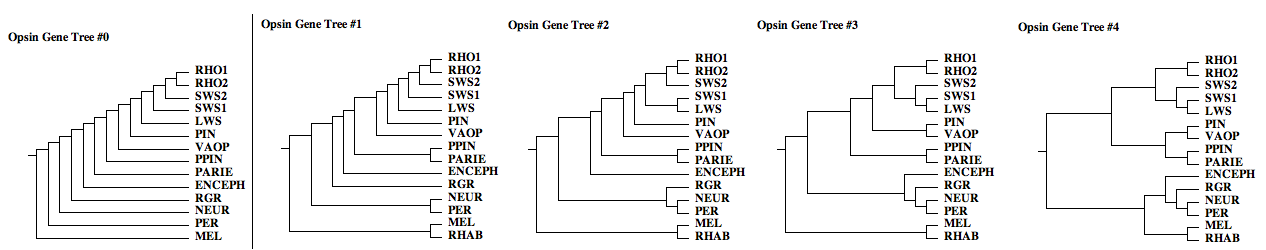
The opsin gene trees below illustrate only a few of the myriad possibilities, even beginning with commonsense ordering (blast nearest neighbors). Because these gene families originated long ago and are only known from remotely related representatives in extant species with wildly differently mutational mechanisms and histories, the true tree cannot be reliably infered from maximal likelihood. (Indeed no two attempts have ever come up with the same gene tree!) Instead, we're going to keep this set of gene trees in view as we analyze the implications of rare genomic events such as indels and intron gains and losses.
On 26 Nov 07, I added 41 new sequences, mostly arthropod rhabdomeric imaging opsins, extracting them from a 2007 pancrustacean opsin paper, using the much-studied accessions in their Table 1, as ordered phylogenetically according to their Fig.3, with subsampling to avoid too-close sequences and narrow lineage-specific expansions.
This involved replacing a few defective accessions and partial sequences with comparable complete ones, favoring sequences with completed or planned genome projects as these can be directly intronated and their synteny determined. Lambda max values were helpfully compiled for all these opsins by the original authors; I have integrated that data as a field in the fasta header database.
This significantly upgrades the resolving power of the Opsin Classifier vis-a-vis these 8 new classes of protostome opsins. This does however raise serious nomenclature issues because of short-sighted nomenclature choices such as rhodopsin or LWS for fruitfly and human genes, which may be vaguely homologous in the distant pre-Bilateran GPCR past but are certainly not orthologous as implied by a common name. The new gene headers are preceded by group name (eg INSE for insect) to disambiguate this in Opsin Classifier output.
Additional ecdysozoan and lophotrochozoan opsins are needed, not just the well-characterized annelid sequences from Platynereis but whatever new that can be extracted from invertebrate genome projects; some of these are ciliary and conversely some deuterostome opsins are non-ciliary. Melanopsin/enchepalopsin appear at the heart of the Big Switchover that took place in chordates -- their imaging opsins did not arise from gene duplication and divergence of anything we see among protostomal imaging opsins (including any reconstructed ancestor).
n fact, none of the opsin genes in Urbilatera destined to become rhabdomeric imaging opsins in living arthropods (even all of protostomia) seems to have descended to any deuterostome. It may turn out that none of the opsin genes in Ur-Bilatera destined to become ciliary imaging opsins in living vertebrates (even all of deuterostomia) survived in any protostome. The pool of GPCR genes was already large and signalling diversified. However lophotrochozoa and basal ecdysozoan ciliary opsins are still largely unexplored.
Worse, a similarly 'bad Venn diagram' could hold in Ur-eumetazoa. Here though the only two cnidarians with sequence data (Hydra and Nematostella) were probably not the best choices for finding opsins. Hence the recommendation to sequence a full-featured cubomedusan.
Please do not add or edit sequences at this time -- email me instead. tom @ cyber-dyne. com (no spaces). After upgrading the Cnidarian and Protostome opsin content, I will refresh alignments, fasta headers, add sections on rare genomic event sectors (indels and introns), provide some ancestral sequences at the common ancestor to lamprey, and post a definitive gene tree.
The 208 sequences below are now organized into deuterostomes, lophotrochozoans, and ecdysozoan divisions broken refined into ciliary, rhabdomeric, or neither. Even with the full set copied into the Opsin Classifier, results are obtained in 6 seconds just using a conventional DSL internet connection.
>RHO1_homSap Homo sapiens (human) Gt 0...2.1.0.0 indel -MBD4 +IFT122 +H1FOO -PLXND1 349 aa 497 nm 16565402 NM_000539 rod rhodopsin RHO ciliary all GT-AG 0 MNGTEGPNFYVPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLEGFFATLG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSR 2 1 YIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQ 0 0 FRNCMLTTICCGKNPLGDDEASATVSKTETSQVAPA* 0 >RHO1_monDom Monodelphis domesticus (opossum) Gt 0...2.1.0.0 indel -MBD4 +IFT122 +H1FOO -PLXND1 349 aa 000 nm no_ref genome rod rhodopsin 0 MNGTEGPNFYVPFSNKTGTVRSPFEEPQYYLADPWQFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAIADLFMVFGGFTMTLYTSLHGYFVFGPTGCNLEGFFATLG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIIGVAFTWVMALACAFPPLIGWSR 2 1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSNFGPIFMTIPAFFAKSSSVYNPVIYIMMNKQ 0 0 FRTCMITTLCCGKNPLGDDEASATASKTETSQVAPA* 0 >RHO1_ornAna Ornithorhynchus anatinus (platypus) Gt 0...2.1.0.0 indel - +IFT122 - -PLXND1 354 aa 000 nm ABN43074 17339011 rod rhodopsin 0 MNGTEGQDFYIPMSNKTGVVRSPFEYPQYYLAEPWQYSVLAAYMFMLIMLGFPINFLTLYVTIQHKKLRTPLNYILLNLAFANHFMVLGGFTTTLYTSLHGYFVFGPTGCNIEGFFATLG 1 2 GEIALWSLVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWIMALACALPPLVGWSR 2 1 YIPEGMQCSCGIDYYTLRPEVNNESFVIYMFVVHFTIPMTIIFFCYGRLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTVPAFFAKSSAIYNPVIYIMMNKQ 0 0 FRNCMLTTICCGKNPLGDDEASATASKTEQSSVSTSQVSPA* 0 >RHO1_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel -MBD4 +IFT122 +H1FOO -PLXND1 352 aa 000 nm 1385866 NM_205490 rod rhodopsin RH1 0 MNGTEGQDFYVPMSNKTGVVRSPFEYPQYYLAEPWKFSALAAYMFMLILLGFPVNFLTLYVTIQHKKLRTPLNYILLNLVVADLFMVFGGFTTTMYTSMNGYFVFGVTGCYIEGFFATLG 1 2 GEIALWSLVVLAVERYVVVCKPMSNFRFGENHAIMGVAFSWIMAMACAAPPLFGWSR 2 1 YIPEGMQCSCGIDYYTLKPEINNESFVIYMFVVHFMIPLAVIFFCYGNLVCTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTNQGSDFGPIFMTIPAFFAKSSAIYNPVIYIVMNKQ 0 0 FRNCMITTLCCGKNPLGDEDTSAGKTETSSVSTSQVSPA* 0 >RHO1_anoCar Anolis carolinensis (lizard) Gt 0...2.1.0.0 indel -MBD4 +IFT122 - -PLXND1 353 aa 000 nm no_ref genome rod rhodopsin 0 MNGTEGQNFYVPMSNKTGVVRNPFEYPQYYLADPWQFSALAAYMFLLILLGFPINFLTLFVTIQHKKLRTPLNYILLNLAVANLFMVLMGFTTTMYTSMNGYFIFGTVGCNIEGFFATLG 2 2 GEMGLWSLVVLAVERYVVICKPMSNFRFGETHALIGVSCTWIMALACAGPPLLGWSR 2 1 YIPEGMQCSCGVDYYTPTPEVHNESFVIYMFLVHFVTPLTIIFFCYGRLVCTVKA 0 0 AAAQQQESATTQKAEREVTRMVVIMVISFLVCWVPYASVAFYIFTHQGSDFGPVFMTIPAFFAKSSAIYNPVIYILMNKQ 0 0 FRNCMIMTLCCGKNPLGDEDTSAGTKTETSTVSTSQVSPA* 0 >RHO1_xenTro Xenopus tropicalis (frog) Gt 0...2.1.0.0 indel -MBD4 +IFT122 - -PLXND1 355 aa 000 nm no_ref genome rod rhodopsin 0 MNGTEGPNFYIPMSNKTGVVRSPFDYPQYYLAEPWKYSALAAYMFLLILLGFPINFMTLYVTIQHKKLRTPLNYILLNLVFANHFMVLCGFTVTMYTSMHGYFIFGQTGCYIEGFFATLG 1 2 GEMALWSLVVLAIERYVVVCKPMANFRFGENHAIMGVVFTWIMALSCAAPPLFGWSR 2 1 YIPEGMQCSCGVDYYTLKPEVNNESFVVYMFIVHFTIPLCVIFFCYGRLLCTVKE 0 0 AAAQQQESATTQKAEKEVTRMVVMMVIFFLICWVPYAYVAFYIFTHQGSDFGPVFMTVPAFFAKSSAIYNPVIYIVLNKQ 0 0 FRNCLITTLCCGKNPFGDEEGSSAASSKTEASSVSSSQVSPA* 0 >RHO1_neoFor Neoceratodus forsteri (lungfish) Gt 0...2.1.0.0 indel x x x x 355 aa 000 nm 17961206 EF526299 rod rhodopsin 0 MNGTEGPNFYVPMTNKTGVVRSPFEYPQYYLADPWKYSALAAYMFFLILTGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTMYTAMNGYFVFGVVGCNLEGFFATFG 1 2 GIIALWCLVVLAIERYIVVCKPISNFRFGENHAIMGVVFTWIMALACAGPPLFGWSR 2 1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFIVHFTIPLIIIFFCYGRLMCTVKE 0 0 AAAQQQESATTQKAEKEVTRMVYIMVISYLVCWLPYASVSFYIFTHQGSDFGPVFMTVPAFFAKTASVYNPVIYILMNKQ 0 0 FRNCMITTLCCGKNPFGDEETTSAGTSKTEASSVSSSQVSPA* 0 >RHO1_latCha Latimeria chalumnae (coelacanth) Gt 0...2.1.0.0 indel x x x x 354 aa 478 nm 10339578 AAD30519 rod rhodopsin 0 MNGTEGPNFYVPMSNKTGVVRNPFEYPQYYLADPWKYSALAAYMFFLILVGFPINFLTLFVTIQHKKLRTPLNYILLDLAVADLCMVFGGFFVTMYSSMNGYFVLGPTGCNIEGFFATLG 1 2 GQVALWALVVLAIERYVVVCKPMSNFRFGENHAIMGVIFTWIMALSCAVPPLFGWSR 2 1 YIPEGMQSSCGVDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGRLVCTVKD 0 0 AAAQQQESATTQKAEKEVTRMVIVMVISFLVCWVPYASVAAYIFFNQGSEFGPVFMTAPSFFAKSASFYNPVIYILLNKQ 0 0 FRNCMITTLCCGKNPFGDEDATSAAGSSKTEASSVSSSSVSPA* 0 >RHO1_takRub Takifugu rubripes (pufferfish) Gt 0...2.1.0.0 indel -MBD4 +IFT122 - -PLXND1 355 aa 000 nm 12783465 AF201472 rod rhodopsin 0 MNGTEGPNFYIPMSNKTGVVRSPFEYPQYYLAEPWKYSLVAAYMLFLIITAFPVNFLTLFVTVKHKKLRTPLNYVLLNLAVADLFMVIGGFTVTLYTALHAYFVLGVTGCNIEGFFATLG 1 2 GEIALWSLVVLAVERYIVVCKPMTNFRFGEKHAIAGLVFTWIMALTCATPPLLGWSR 2 1 YIPEGMQCSCGIDYYTPKPEINNTSFVIYMFILHFSIPLAIIFFCYSRLLCTVRA 0 0 AAALQQESETTQRAEKEVTRMVIVMVISFLVCWVPYASVAWYIFANQGTEFGPVFMTAPAFFAKSAALYNPVIYILLNRQ 0 0 FRNCMITTVCCGKNPFGDDDAATTVSKTQSSSVSSSQVAPA* 0 >RHO1_leuEri Leucoraja erinacea (skate) Gt 0...2.1.0.0 indel x x x x 355 aa 000 nm 9256070 U81514 rod rhodopsin 0 MNGTEGENFYVPMSNKTGVVRSPFDYPQYYLGEPWMFSALAAYMFFLILTGLPVNFLTLFVTIQHKKLRQPLNYILLNLAVSDLFMVFGGFTTTIITSMNGYFIFGPAGCNFEGFFATLG 1 2 GEVGLWCLVVLAIERYMVVCKPMANFRFGSQHAIIGVVFTWIMALSCAGPPLVGWSR 2 1 YIPEGLQCSCGVDYYTMKPEVNNESFVIYMFVVHFTIPLIVIFFCYGRLVCTVKE 0 0 AAAQQQESESTQRAEREVTRMVIIMVVAFLICWVPYASVAFYIFINQGCDFTPFFMTVPAFFAKSSAVYNPLIYILMNKQ 0 0 FRNCMITTICLGKNPFEEEESTSASASKTEASSVSSSQVAPA* 0 >RHO1_calMil Callorhinchus milii (elephantfish) Gt 0...2.1.0.0 indel x x x x 355 aa 000 nm no_ref genome rod rhodopsin complete wgs 0 MNGTEGENFYIPMSNKTGVVRSPFEYPQYYLAEPWQFSILAAYMFFLIITCFPVNFLTLYVTFEHKKLRQPLNFILLNLAVADLFMVFGGFFITVYTSLHGYFVFGVTGCNFEGFFATLG 1 2 GEIGLWSLVVLAIERYVVVCKPMSNFRFGTNHAIMGVAFTWVMALACAVPPLMGWSR 2 1 YIPEGLQCSCGVDYYTLKPEINNESFVIYMFVVHFLIPLIIIFFCYGRLVCTVKE 0 0 AAAQQQESESTQRAEREVTRMVIIMVIFFLICWVPYASVAFFIFTNQGSEFGPIFMAVPAFFAKSSALYNPLIYILLNKQ 0 0 FRNCMITTLCCGKNPFEEDESTSAAASKTEASSVSSSQVSPA* 0 >RHO1_petMar Petromyzon marinus (lamprey) Gt 0...2.1.0.0 indel x x x x 354 aa 000 nm no_ref genome rod rhodopsin 0 MNGTEGENFYIPFSNKTGLARSPFEYPQYYLAEPWKYSVLAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAVANLFMVLFGFTLTMYSSMNGYFVFGPTMCNFEGFFATLG 1 2 GEMSLWSLVVLAIERYIVICKPMGNFRFGSTHAYMGVAFTWFMALSCAAPPLVGWSR 2 1 YLPEGMQCSCGPDYYTLNPNFNNESFVIYMFLVHFIIPFIVIFFCYGRLLCTVKE 0 0 AAAAQQESASTQKAEKEVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTVPAFFAKTSALYNPIIYILMNKQ 0 0 FRNCMITTLCCGKNPLGDEDSGASTSKTEVSSVSTSQVSPA* 0 >RHO1_geoAus Geotria australis (lamprey) Gt 0...2.1.0.0 indel x x x x 354 aa 497 nm 17463225 AY366493 rod rhodopsin rodRhA 0 MNGTEGQNFYIPFSNKTDVARSPFEYPQYYLAEPWKFSALAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAVSNLFMILFGFTTTMYTSMNGYFVFGPTMCSIEGFFATLG 1 2 GEVSLWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVALTWVMALSCAAPPLLGWSR 2 1 YLPEGMQCSCGPDYYTMNPTYNNESFVIYMFIVHFTIPFVIIFFSYGRLLCTVKE 0 0 AAAAQQESASTQKAEKEVTRMVVLMVVGFLVCWVPYASVAFYIFTNQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ 0 0 FRNCMITTLCCGKNPLGDDDSGASTSKTEVSSVSTSQVAPA* 0 >RHO1_letJap Lethenteron japonicum (lamprey) Gt 0...2.1.0.0 indel x x x x 354 aa 000 nm 15096614 AB116382 cone rhodopsin 0 MNGTEGDNFYVPFSNKTGLARSPYEYPQYYLAEPWKYSALAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAMANLFMVLFGFTVTMYTSMNGYFVFGPTMCSIEGFFATLG 1 2 GEVALWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVAFTWIMALACAAPPLVGWSR 2 1 YIPEGMQCSCGPDYYTLNPNFNNESYVVYMFVVHFLVPFVIIFFCYGRLLCTVKE 0 0 AAAAQQESASTQKAEKEVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ 0 0 FRNCMITTLCCGKNPLGDDESGASTSKTEVSSVSTSQVSPA* 0 >RHO2_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel -IHPK3 -LEMD2 -GRM4 +HMGA1 356 aa 000 nm 2268324 NP_990771 cone rhodopsin 0 MNGTEGINFYVPMSNKTGVVRSPFEYPQYYLAEPWKYRLVCCYIFFLISTGLPINLLTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFVFGPVGCAVEGFFATLG 1 2 GQVALWSLVVLAIERYIVVCKPMGNFRFSATHAMMGIAFTWVMAFSCAAPPLFGWSR 2 1 YMPEGMQCSCGPDYYTHNPDYHNESYVLYMFVIHFIIPVVVIFFSYGRLICKVRE 0 0 AAAQQQESATTQKAEKEVTRMVILMVLGFMLAWTPYAVVAFWIFTNKGADFTATLMAVPAFFSKSSSLYNPIIYVLMNKQ 0 0 FRNCMITTICCGKNPFGDEDVSSTVSQSKTEVSSVSSSQVSPA* 0 >RHO2_anoCar Anolis carolinensis (lizard) Gt 0...2.1.0.0 indel -IHPK3 -LEMD2 -GRM4 +HMGA1 356 aa 000 nm no_ref genome cone rhodopsin 0 MNGTEGINFYVPLSNKTGLVRSPFEYPQYYLAEPWKYKVVCCYIFFLIFTGLPINILTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFIFGPIGCAIEGFFATLG 1 2 GQVALWSLVVLAIERYIVVCKPMGNFRFSATHALMGISFTWFMSFSCAAPPLLGWSR 2 1 YIPEGMQCSCGPDYYTLNPDYHNESYVLYMFGVHFVIPVVVIFFSYGRLICKVRE 0 0 AAAQQQESASTQKAEREVTRMVILMVLGFLLAWTPYAMVAFWIFTNKGVDFSATLMSVPAFFSKSSSLYNPIIYVLMNKQ 0 0 FRNCMITTICCGKNPFGDEDVSSSVSQSKTEVSSVSSSQVSPA* 0 >RHO2_gekGek Gekko gekko (gecko) Gt 0...2.1.0.0 indel x x x x 356 aa 000 nm 11591478 AY024356 cone rhodopsin in pure rod-retina 0 MNGTEGINFYVPLSNKTGLVRSPFEYPQYYLADPWKFKVLSFYMFFLIAAGMPLNGLTLFVTFQHKKLRQPLNYILVNLAAANLVTVCCGFTVTFYASWYAYFVFGPIGCAIEGFFATIG 1 2 GQVALWSLVVLAIERYIVICKPMGNFRFSATHAIMGIAFTWFMALACAGPPLFGWSR 2 1 FIPEGMQCSCGPDYYTLNPDFHNESYVIYMFIVHFTVPMVVIFFSYGRLVCKVRE 0 0 AAAQQQESATTQKAEKEVTRMVILMVLGFLLAWTPYAATAIWIFTNRGAAFSVTFMTIPAFFSKSSSIYNPIIYVLLNKQ 0 0 FRNCMVTTICCGKNPFGDEDVSSSVSQSKTEVSSVSSSQVAPA* 0 >RHO2_neoFor Neoceratodus forsteri (lungfish) Gt 0...2.1.0.0 indel x x x x 356 aa 000 nm 17961206 EF526299 cone rhodopsin 0 MNGTEGINFYVPHSNKTGVVRSPFEYPQYYLADPWKYSIVCAYMFFLIITGLPINLLTLVVTFKHKKLRQPLNYILVNLAVADLFMVCFGFTVTFSTAINGYFIFGPRGCAIEGFMATLG 1 2 GEVALWSLVVLAIERYIVVCKPMGNFRFSNNHSIIGIVFTWLAALSCAAPPLFGWSR 2 1 YLPEGMQCSCGPDYYTMNPDYHNESFVIYMFVVHFFIPVIVIFVSYGRLICKVKE 0 0 AAAQQQESASTQKAEREVTRMVILMVIGFMTAWTPYATVAFWIFMNKGAEFGATFMAAPAFFSKSSALYNPIIYVLMNKQ 0 0 FRNCMVTTLCCGKNPFGDDDVSSSVSAGKTEVSSVSSSQVSPA* 0 >RHO2_latCha Latimeria chalumnae (coelacanth) Gt 0...2.1.0.0 indel x x x x 355 aa 485 nm 10339578 AH007713 cone rhodopsin RH2 0 MNGTEGMNFYVPLSNRTGLVRSPFEYTQYYLAEPWKFSVLCAYMFLLIILGFPINFLTLLVTFKHKKLRQPLNYILVNLAVASLFMVVFGFTVTFYSSLNGYFVLGPMGCAMEGFFATLG 1 2 GQVALWSLVVLAIERYIVVCKPMGNFRFASSHAIMGIAFTWIMALACAAPPLVGWSR 2 1 YIPEGLQCSCGPDYYTLNPDFHNESYVMYLFLVHFLLPIIIIFFTYGRLICKVKE 0 0 AAAQQQESASTQKAEKEVTRMVILMVIGFLTAWVPYASAAFWIFCNRGAEFTATLMTVPAFFSKSSCLFNPIIYVLLNKQ 0 0 FRNCMITTLCCGKNPLGDDDTSSAVSQSKTDVSSVSSSQVSPA* 0 >RHO2_geoAus Geotria australis (lamprey) Gt 0...2.1.0.0 indel x x x x 355 aa 492 nm 17463225 AY366494 cone rhodopsin RhB no petMar 0 MNGTEGANFYIPFHNRTGVVRSPYEYPQYYLADPWMYSAISAYVFTLILIGFPVNFMTLFVTFKLKKLRQPLNFILVNLCVADLLMIMFGFTTTFYTAMNGYFVFGPTGCNIEGFFATLG 1 2 GEVSLWSLVMLAIERYIVVCKPMGNFRFATTHAALGVVFTWVMASACAVPPLVGWSR 2 1 YIPEGMQCSCGPDYYTLNPKYYNESYVIYLFLVHFLLPVTIIFFTYGRLICTVKE 0 0 AAAQQQESASTQKAEREVTRMVIIMVVGFLVCWVPYASFAFYLFMNKGILFSATAMTVPAFFSKSSVLYNPIIYVLLNKQ 0 0 FRTCMVTTLFCGKNPFGEDDSSMVSTSKTEVSSVSSSQVSPS* 0 >SWS2_ornAna Ornithorhynchus anatinus (platypus) Gt 0...2.1.0.0 indel -IRAK1 -MECP2 - +TKTL1 364 aa 000 nm 17339011 ABN43074 cone short blue tandem -FLNB--+MECP2 with MWS1 0 MHKTHRNLQNELPEDFFIPLPLDTDNITSLSPFLVPQTHLGGSGIFMSLAAFMFLLITLGFPINLLTVICTIKYKKLRSHLNYILVNLAVSNMLVVCVGSATAFYSFAHMYFVLGPTACKIEGFAATLG 1 2 GMVSLWSLAVIAFERFLVICKPLGNLSFRGTHAIFGCAATWVFGLAASLPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTNNKWNNESYVIFLFSFCFGVPLSIIIFSYGRLLLTLRA 0 0 VAKQQEQSATTQKAEREVTKMVIVMVLGFLVCWLPYASFSLWVVTNRGQVFDLRMASIPSVFSKASTIYNPIIYVFMNKQ 0 0 FRSCMLKLVFCGKSPFGDEDEISGSSQATQVSSVSSSQVSPA* 0 >SWS2_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel x x x x 362 aa 000 nm 7975342 NP_990848 cone short2 blue 0 MHPPRPTTDLPEDFYIPMALDAPNITALSPFLVPQTHLGSPGLFRAMAAFMFLLIALGVPINTLTIFCTARFRKLRSHLNYILVNLALANLLVILVGSTTACYSFSQMYFALGPTACKIEGFAATLG 1 2 GMVSLWSLAVVAFERFLVICKPLGNFTFRGSHAVLGCVATWVLGFVASAPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTDNKWHNESYVLFLFTFCFGVPLAIIVFSYGRLLITLRA 0 0 VARQQEQSATTQKADREVTKMVVVMVLGFLVCWAPYTAFALWVVTHRGRSFEVGLASIPSVFSKSSTVYNPVIYVLMNKQ 0 0 FRSCMLKLLFCGRSPFGDDEDVSGSSQATQVSSVSSSHVAPA* 0 >SWS2_taeGut Taeniopygia guttata (finch) Gt 0...2.1.0.0 indel x x x x 363 aa 000 nm no_ref genome cone short2 0 MPKPREMRDELPEDFYIPMSLETPNLTALSPFLVPQTHLGSPGIFKAMAAFMFLLVLLGVPINALTVLCTAKYKKLRSHLNYILVNLAVANLLVVCVGSTTAFYSFSQMYFALGPLACKIEGFTATLG 1 2 GMVSLWSLAVVAFERFLVICKPLGNFTFRGSHAVLGCAITWIFGLIASLPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTDNKWNNESYVIFLFCFCFGFPLTVIVFSYGRLLLTLRA 0 0 VAKQQEQSASTQKAEREVTKMVVVMVLGFLVCWLPYCSFALWVVTHRGHPFDLGLASIPSVFSKASTVYNPIIYVFMNKQ 0 0 FRSCMLKLVFCGRSPFGDEDDVSGSSQATQVSSVSSSQVSPA* 0 >SWS2_utaSta Uta stansburiana (lizard) Gt 0...2.1.0.0 indel x x x x 364 aa 000 nm 16543463 DQ100326 cone short 0 MHNSRPHSRDDLPEDFFIPMPLDVANITTLSPFLVPQTHLGSPALFMGMAAFMFLLIILGVPINVLTIFCTFKYKKLRSHLNYILVNLAVSNLLVVCIGSTTAFYSFAQMYFSLGPTACKIEGFAATLG 1 2 GMVSLWSLAVVAFERFLVICKPLGNFSFRGTHAIIGCIITWVFGLVASLPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTNNKWNNESYVLFLFSFCFGVPLSVIIFSYGRLLLTLRA 0 0 VAKQQEQSATTQKAEREVTKMVVVMVMGFLVCWLPYASFALWVVTHRGEPFDVRLATIPSVFSKASSVYNPVIYVFMNKQ 0 0 FRSCMLKLVFCGKSPFGDEDDVSGSSQTTQVSSVSSSQVSPA* 0 >SWS2_xenTro Xenopus tropicalis (frog) Gt 0...2.1.0.0 indel -IRAK1 -MECP2 - - 363 aa 000 nm no_ref genome cone short 0 MSKGRPDLRMEMPDEFYVPIPLETTNISSLSPFLVPQTHLGTPGIFMSISAFMLFTIIFGFPLNLLTIICTVKYKKLRSHLNYILVNLAVANLIVICFGSTTAFYSFSQMYFSLGTLACKIEGFTATLG 1 2 GIIGLWSLAVVAFERFLVICKPMGNFTFRESHAVLGCILTWVIGLVAAIPPLLGWSR 2 1 YIPEGLQCSCGPDWYTVNNKWNNESYVLFLFCFCFGFPLAIIVFSYGRLLLALHA 0 0 VAKQQEQSATTQKAEREVTRMVIVMVVGFLVCWLPYASFALWAVTHRGELFDLRMSSVPSVFSKASTVYNPFIYIFMNRQ 0 0 FRSCMMKMIFCGKNPLGDDEETSVSGSTQVSSVSSSQIAPS* 0 >SWS2_neoFor Neoceratodus forsteri (lungfish) Gt 0...2.1.0.0 indel x x x x 364 aa 000 nm 17961206 EF526299 cone short 0 MHRTKPDPQEDLPDDFYIPVSLNTNNITMLSPFLVPQTHLGSPSVFMVLSVFMFFLLITGIPINVLTIICTFKYKKLRSHLNYILVNLAVANLIVVGFGSTTAFYSFSQMYFAWGPLACKIEGFAATLG 1 2 GMVSLWSLAVVAFERFLVICKPLGNFTFRSTHAIIGCVATWVFGLISSAPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTNNKWNNESYVIFLFCFCFGFPLSVIIFSYGRLLMTLRA 0 0 VAKQQEQSASTQKAEREVTKMVVVMVLGFLVCWLPYTVFSLWVVTHRGESFELALGSIPAVFSKSSTVYNPLIYVFMNKQ 0 0 FRSCMMKLIFCGKSPFGDEDDASSASQSTQVSSVSSSQVAPA* 0 >SWS2_takRub Takifugu rubripes (pufferfish) Gt 0...2.1.0.0 indel x x x x 351 aa 000 nm no_ref genome cone short2 0 MRGVRQHEFQEDFYIPIPLDVDNITALSPFLVPQDHLGSPAVFYGMSAFMFFLFVAGTGINVLTIACTIQYKKLRSHLNYILVNLAFSNLLVTTVGSFTCFCCFFVRYMIVGPLGCKIEGFAATLG 1 2 GMVSLWSLAVVAFERWLVVCKPLGNFIFKPDHAIVCCIFTWFFALIISAPPLFGWSR 2 1 YIPEGFQCSCGPDWYTTGNKYNNESYVWFIFGFGFAVPLFVIVFCYSQLLVMLKS 0 0 AKAQAESASTQKAEREVTRMVVVMILGFLVCWLPYASFALWVVNNRGTPFDLRLATIPACFSKASTVYNPIIYVVLNKQ 0 0 FRSCMKKMLGMSGGDDEESSSQSVTEVSKVSPS* 0 >SWS2_gasAcu Gasterosteus aculeatus (stickleback) Gt 0.2.2.1.0.0 indel x x x x 359 aa 000 nm no_ref genome cone short 0 MKHGRVPEIPEDFYIPISLDTDNITSLSPFLVPQDHLASKATFYSLAFYMFFILIVGTFINALTVACTVQNKKLRSHLNYILVNLAVSNLLVSGVGAFTAFLSFAARYFVLGTLACKVEGFLATLG 1 2 GMVSLWSLAVIAFERWLVICKPLGNFIFKPDHALVCCAFTWVFALAASAPPLVGWSR 2 1 YIPEGLQCSCGPDWYTTNNKYNNESYVLFLFGFCFAVPFCTICFCYSQLLFTMKMA 0 0 AKAQAESASTQKAEREVTRMVVLMVMGFLVCWMPYASFALWVVNNRGQTFDLRFASIPSVFSKSSAVYNPVIYVLLNKQ 0 0 FRSCMMKMLGMGGGDDEESSTSSVTEVSKVGPA* 0 >SWS2_geoAus Geotria australis (lamprey) Gt 0...2.1.0.0 indel x x x x 362 aa 439 nm 17463225 AY366492 cone short2 blue retinal petMar ps 0 MYQGKSTQVDDLPEDFYIPIALNVKNMSELSPFLVPQVHLGDSFIFYGMSAFMLFLVLAGFPLNFLTVFVTIKYKKLRSHLNYILVNLAIANLIVVCCGSTLAFYSFMHKYFILGPLFCKMEGFTATLG 1 2 GMLSLWSLAVLAFERCLVICKPFGNIAFRGTHALIRCGFAWAAAIAASTPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTNNKYNNESYVMFLFIFCFGTPFTIIIVSYSKLILTLRA 0 0 AAAQQQESASTQKAEKEVSRMVVIMVGGFLVCWLPYASLALWIVFNRGSPFDLRLATIPSVFSKASTVYNPVIYIFLNKQ 0 0 FRSCMMKTIFCGKNPLGDDEDATSTTTQVSSVSTSQVAPA* 0 >SWS1_homSap Homo sapiens (human) Gt 0.2.2.1.0.0 indel -FAM137A -CALU -NAG6 -FLNC 348 aa 000 nm 1385866 NP_990769 cone short 0 MRKMSEEEFYLFKNISSVGPWDGPQYHIAPVWAFYLQAAFMGTVFLIGFPLNAMVLVATLRYKKLRQPLNYILVNVSFGGFLLCIFSVFPVFVASCNGYFVFGRHVCALEGFLGTVA 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSKHALTVVLATWTIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHGLDLRLVTIPSFFSKSACIYNPIIYCFMNKQ 0 0 FQACIMKMVCGKAMTDESDTCSSQKTEVSTVSSTQVGPN* 0 >SWS1_monDom Monodelphis domesticus (opossum) Gt 0...2.1.0.0 indel -FAM137A -CALU -NAG6 -FLNC 347 aa 000 nm no_ref genome cone short 0 MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHFQTVFMGFVFCAGTPLNAVVLVATLRYKKLRQPLNYILVNVSLCGFIFCIFAVFTVFISSSQGYFIFGRHVCAMEAFLGSVA 1 2 GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWVIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIMPLFLICFSYSQLLRALRA 0 0 VAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNQNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FHACIMEMVCRKPMTDDSDVSSSQKTEVSAVSSSQVGPT* 0 >SWS1_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel x x x x 348 aa 000 nm no_ref genome cone short1 violet 0 MSSDDDFYLFTNGSVPGPWDGPQYHIAPPWAFYLQTAFMGIVFAVGTPLNAVVLWVTVRYKRLRQPLNYILVNISASGFVSCVLSVFVVFVASARGYFVFGKRVCELEAFVGTHG 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSRHALLVVVATWLIGVGVGLPPFFGWSR 2 1 YMPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FRACIMETVCGKPLTDDSDASTSAQRTEVSSVSSSQVGPT* 0 >SWS1_taeGut Taeniopygia guttata (finch) Gt 0...2.1.0.0 indel x x x x 347 aa 000 nm no_ref genome cone short1 0 MDEEEFYLFKNQSSVGPWDGPQYHIAPMWAFYLQTIFMGLVFVAGTPLNAIVLIVTIKYKKLRQPLNYILVNISVSGLMCCVFCIFTVFIASSQGYFVFGKHMCAFEGFAGATG 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSRHALLVVAATWIIGVGVAIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCMCYVPYAALAMYMVNNREHGIDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0 0 FRACIMETVCGRPMTDDSEVSSSAQRTEVSSVSSSQVGPS* 0 >SWS1_anoCar Anolis carolinensis (lizard) Gt 0.2.2.1.0.0 indel - -CALU - - 347 aa 000 nm no_ref genome cone short 0 MSGQEDFYLFENISSVGPWDGPQYHIAPMWAFYFQTAFMGFVFFAGTPLNAIILIVTVKYKKLRQPLNYILVNISFAGFLFCTFSVFTVFMASSQGYFFFGRHVCAMEAFLGSVA 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSRHALLVVAATWIIGVGVAIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLTLIIFSYSQLLGALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYASLAMYMVNNRDHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0 0 FRACILETVCGKPMSDESDVSSSAQKTEVSSVSSSQVSPS* 0 >SWS1_utaSta Uta stansburiana (lizard) Gt 0...2.1.0.0 indel x x x x 348 aa 000 nm 16543463 DQ100325 cone short 0 MSGEEDFYLFENISSVGPWDGPQYHIAPMWAFYFQTAFMGFVFFAGTPLNAIILIVTVKYKKLRQPLNYILVNISFAGFLFCVFSVFTVFLASSQGYFFFGRHICALEAFLGSVA 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSKHALLVVAATWFIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLTLIIFSYSQLLGALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGIDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FRACIMETVCGKPMTDESDVSSSAQKTEVSSVSSSQVSPS* 0 >SWS1_xenLae Xenopus laevis (frog) Gt 0...2.1.0.0 indel - -CALU - - 348 aa 000 nm no_ref genome cone short 0 MLEEEDFYLFKNVSNVSPFDGPQYHIAPKWAFTLQAIFMGMVFLIGTPLNFIVLLVTIKYKKLRQPLNYILVNITVGGFLMCIFSIFPVFVSSSQGYFFFGRIACSIDAFVGTLT 1 2 GLVTGWSLAFLAFERYIVICKPMGNFNFSSSHALAVVICTWIIGIVVSVPPFLGWSR 2 1 YMPEGLQCSCGPDWYTVGTKYRSEYYTWFIFIFCFVIPLSLICFSYGRLLGALRA 0 0 VAAQQQESASTQKAEREVSRMVIFMVGSFCLCYVPYAAMAMYMVTNRNHGLDLRLVTIPAFFSKSSCVYNPIIYSFMNKQ 0 0 FRGCIMETVCGRPMSDDSSVSSTSQRTEVSTVSSSQVSPA* 0 >SWS1_neoFor Neoceratodus forsteri (lungfish) Gt 0...2.1.0.0 indel x x x x 347 aa 000 nm 17961206 EF526299 cone short 0 MSGEEEFYLFKNISSVGPWDGPQYHIAPKWAFFLQAAFMGFVLFVGTPLNAIVLFVTIKYKKLQQPLNYILVNISLAGFIFCFFGVFAVFIASCQGYFIFGKTVCALEGFTGSVA 1 2 GLVTGWSLAILAFERYLVICKPIGNFRFGSKHSMIAVVAAWVIGVGVSIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIIPLFIICFSYSQLLGALRA 0 0 VAAQQQESATTQKAEREVSRMIIVMVGSFCVCYVPYAALAMYMVNNRDHGIDLRLVTIPAFFSKSSFVYNPIIYCFMNKQ 0 0 FRACIMQTVFGKPMTDDSDISSSGKTEVSSVSSSQVNPS* >SWS1_danRer Danio rerio (zebrafish) Gt 0...2.1.0.0 indel - -CALU - - 337 aa 000 nm no_ref genome cone short1 0 MDAWAVQFGNASKVSPFEGEQYHIAPKWAFYLQAAFMGFVFIVGTPMNGIVLFVTMKYKKLRQPLNYILVNISLAGFIFDTFSVSQVSVCAARGYYSLGYTLCSMEAAMGSIA 1 2 GLVTGWSLAVLAFERYVVICKPFGSFKFGQGQAVGAVVFTWIIGTACATPPFFGWSR 2 1 YIPEGLGTACGPDWYTKSEEYNSESYTYFLLITCFMMPMTIIIFSYSQLLGALRA 0 0 VAAQQAESESTQKAEREVSRMVVVMVGSFVLCYAPYAVTAMYFANSDEPNKDYRLVAIPAFFSKSSSVYNPLIYAFMNKQ 0 0 FNACIMETVFGKKIDESSEVSSKTETSSVSA* 0 >SWS1_oryLat Oryzias latipes (medaka) Gt 0...2.1.0.0 indel - - - - 336 aa 000 nm no_ref genome cone short1 0 MGKYFYLYENISKVGPYDGPQYYLAPTWAFYLQAAFMGFVFFVGTPLNFVVLLATAKYKKLRVPLNYILVNITFAGFIFVTFSVSQVFLASVRGYYFFGQTLCALEAAVGAVA 1 2 GLVTSWSLAVLSFERYLVICKPFGAFKFGSNHALAAVIFTWFMGVGCACPPFFGWSR 2 1 YIPEGLGCSCGPDWYTNCEEFSCASYSKFLLVTCFICPITIIIFSYSQLLGALRA 0 0 VAAQQAESASTQKAEKEVSRMIIVMVASFVTCYGPYALTAQYYAYSQDENKDYRLVTIPAFFSKSSCVYNPLIYAFMNKQ 0 0 FNGCIMEMVFGKKMEEASEVSSKTEVSTDS*0 >SWS1_geoAus Geotria australis (lamprey) Gt 0...2.1.0.0 indel x x x x 346 aa 359 nm 17463225 AY366495 cone short1 UV retinal 0 MSGDEEFYLFKNISKVGPWDGPQFHIAPKWAFYLQAAFMGFVFICGTPLNAIVLVVTIKYKKLRQPLNYILVNISAAGLVFCLFSISTVFVASMQGYFFLGPTICALEAFFGSLA 1 2 GLVTGWSLAFLAAERYIVICKPFGNFRFGSKHALVAVGLTWMLGLSVALPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYKSEYYTYFLFVFCFVVPLSIIIFSYGSLLGTLRA 0 0 VAAQQQESASTQKAEREVSRMVIMMVASFCTCYVPYAALAVYMVTNRDHNIDLRFVTVPAFFSKASCVYNPLIYSFMNKQ 0 0 FRACILETVCGKPITDESETSSSRTEVSSVSTTQMIPG* 0 >LWS_homSap Homo sapiens (human) Gt 0.2.2.1.0.0 indel -IRAK1 -MECP2 -TEX28 +TKTL1 364 aa 530 nm 12853434 NP_000504 cone long OPN1MW deutan 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_monDom Monodelphis domesticus (opossum) Gt 0.2.2.1.0.0 indel -IRAK1 -MECP2 - +TKTL1 368 aa 000 nm no_ref genome cone long 0 MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYVQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYSFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_ornAna Ornithorhynchus anatinus (platypus) Gt 0.2.2.1.0.0 indel -IRAK1 -MECP2 - - 365 aa 000 nm 17339011 ABN43074 cone long LWS green 0 MTPAWNSGVYAARRRFEDEEDTTRTSVFVYTNSNNTR 1 2 DPFEGPNYHIAPRWAYNVTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIFGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLSIISWERWIVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSSVSPA* 0 >LWS_galGal Gallus gallus (chicken) Gt 0.2.2.1.0.0 indel x x x x 363 aa 000 nm 12716987 NM_205438 cone long green iodopsin missing in assembly 0 MAAWEAAFAARRRHEEEDTTRDSVFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVAASVFTNGLVLVATWKFKKLRHPLNWILVNLAVADLGETVIASTISVINQISGYFILGHPMCVVEGYTVSAC 1 2 GITALWSLAIISWERWFVVCKPFGNIKFDGKLAVAGILFSWLWSCAWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFFPLAIIILCYLQVSLAIRA 0 0 VAAQQKESESTQKAEKEVSRMVVVMIVAYCFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSEVSTSRTEVSSVSNSSVSPA* 0 >LWS_anoCar Anolis carolinensis (lizard) Gt 0.2.2.1.0.0 indel - - -TEX28 +TKTL1 366 aa 000 nm no_ref genome cone long 0 MAGTVTEAWDVAVFAARRRNDEDDTTRDSLFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNITSVWMIFVVIASIFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVIASTISVINQISGYFILGHPMCVLEGYTVSTC 1 2 GISALWSLAVISWERWVVVCKPFGNVKFDAKLAVAGIVFSWVWSAVWTAPPVFGWSR 2 1 YWPHGLKTSCGPDVFSGSDDPGVLSYMIVLMITCCFIPLAVILLCYLQVWLAIRA 0 0 VAAQQKESESTQKAEKEVSRMVVVMIIAYCFCWGPYTVFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSNSSVSPA* 0 >LWS_xenTro Xenopus tropicalis (frog) Gt 0.2.2.1.0.0 indel -IRAK1 -MECP2 - - 370 aa 000 nm no_ref genome cone long 0 MASHWNEAVFAARRRNDDDDTTRSSVFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNISSLWMIFVVLASVFTNGLVLVATLKFKKLRHPLNWILVNMAIADLGETVIASTISVCNQIFGYFVLGHPMCILEGYTVSVC 1 2 GIAALWSLTVIAWERWFVVCKPFGNIKFDGKLAATGIIFSWVWAAGWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMLVLMITCCIIPLAIIVLCYMHVWLTIRQ 0 0 VAQQQKESESTQKAEREVSRMVVVMIIAYIFCWGPYTFFACFAAFNPGYNFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIYQLFGKKVDDGSEVSSTSRTEVSSVSNSSVSPA* 0 >LWS_neoFor Neoceratodus forsteri (lungfish) Gt 0.2.2.1.0.0 indel x x x x 365 aa 000 nm 17961206 EF526299 cone long 0 MAEPWDAVLAARRRHQDEETTRSTIFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVFASCFTNGLVLMATYKFKKLRHPLNWILVNLAIADLGETLIASTISVTNQIFGYFILGHPMCMLEGFTVATC 1 2 GITGLWSLTIIAWERWVVVCKPFGNIKFDGKWAAGGIIFSWVWSAFWCAMPLFGWSR 2 1 FWPHGLKTSCGPDVFSGEDKYGTRSFMIALMITCCIIPLGVIILCYIQVWWAIRT 0 0 VAKQQKESESTQKAEKEVSRMVVVMIFAYCFCWGPYTFMACFGAAYPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIYQLLGKKVDDGSELSSTSKTEVSSVSNSSVSPA* 0 >LWS_takRub Takifugu rubripes (pufferfish) Gt 0...2.1.0.0 indel x x x x 358 aa 000 nm no_ref genome cone long 0 MAEEWGKQSFAARRYHEDTTRGSAFVYTNSNHTR 1 2 DPFEGPNYHIAPRWVYNVATVWMFIVVVLSVFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYTVSTC 1 2 GIAALWSLTIISWERWVVVCKPFGNVKFDAKWATGGIVFSWVWAAVWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCIIPLAIIILCYLAVWLAIRS 0 0 VAMQQKESESTQKAEKEVSRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 0 FRVCIMKLFGKEVDDGSEVSTSKTEVSSVAPA* 0 >LWS_gasAcu Gasterosteus aculeatus (stickleback) Gt 0.2.2.1.0.0 indel - - - - 358 aa 000 nm no_ref genome cone long 0 MAEEWGKQAFAARRYNEDTTRGSMFVYTNSNNTK 1 2 DPFEGPNYHIAPRWVYNLSTLWMFIVVALSVFTNGLVLVATAKFKKLQHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYVVSVC 1 2 GITALWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWIWSAVWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCLIPLAIIILCYLAVWLAIRA 0 0 VAMQQKESESTQKAERDVSRMVVVMIVAYIVCWGPYTTFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 0 FRSCIMQLFGKEVDDGSEVSTsKTEVSSVAPA* 0 >LWS_calMil Callorhinchus milii (elephantfish) Gt 0.2.2.1.0.0 indel x x x x 262 aa 000 nm no_ref genome fragment exon break 2 dPFEGPNYHIAPRWAYNLTSVWMVGVVVASVFTNGLVLVATVRFKKLRHPLNWILVNMALADLGETVLASTVSVANQFFGYFILGHPLCVFEGFVVSLC 1 2 GITALWSLTIIAWERWVVVCKPFGNVKFDGKWAAFGIIFSWVWSIGWCLPPVFGWSR 2 0 AEKEVSRMVVVMVAAFCLCWGPYACFAMFSALNPGYAFHPLVASIPSYFAKSSTIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSTSKTDVSSVSNSSVSPA* 0 >LWS_petMar Petromyzon maritimus (lamprey) Gt 0.2.2.1.0.0 indel x x x x 366 aa 000 nm no_ref genome cone traces key to intron 3 position and gapping 0 MTASWQGAMFAARRRQDDEDTTMESLFRYTNENNTK 1 2 DPFEGPNYHIAPRWVFNLTSVWMIIVVVLSLFSNGLVLVATVKFKKLRHPLNWIIVNLAIADILETIFASTISVCNQVYGYFILGHPMCVFEGYVVSTC 1 2 GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIATILIVFSWVWPASWCSLPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFLPLSIIILCYLQVWLAIHS 0 0 VAQQQKESETTQKAERDVSRMVVVMILAYVFCWGPYTFFACFAAANPGYSFHPIAAALPAYFAKGATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSEVSSSSRTEVSSVSNSSVSPA* 0 >LWS_letJap Lethenteron japonicum (lamprey) Gt 0.2.2.1.0.0 indel x x x x 365 aa 000 nm 15096614 AB116381 cone long 0 MTASWHGAVFAARRRNDDEDTTKDSIFRYTNENNTR 1 2 DPFEGPNYHIAPRWMFNLTSVWMIIVVVLSLFTNGLVLVATMKFKKLRHPLNWILVNLAIADILETIFASTISVCNQVFGYFILGHPMCVFEGYVVSTC 1 2 GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIAIILIVFSWVWPACWCSLPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFLPLSVIILCYLQVWLAIHS 0 0 VAQQQKESETTQKAERDVSRMVVVMILAYIFCWGPYTFFACYAAANPGYAFHPLTAALPAYFAKSATIYNPVIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSEVSSASRTEVSSVSNSSISPA* >LWS_geoAus Geotria australis (lamprey) Gt 0.2.2.1.0.0 indel x x x x 365 aa 560 nm 17463225 AY366491 cone long red retinal 0 MAQSWERAMFAARRRQDEDTTKGDLFRYTNENNTR 1 2 DPFEGPNYHIAPRWMYNLTSFWMIIVVILSLFTNGLVLVATLKFKKLRHPLNWILVNLAIADIGETIFASTVSVVNQIFGYFILGHPLCVFEGFTVSVC 1 2 GITALWSLAIISFERWMVVCKPFGNLKFDGKVAIVLIIFSWAWSAGWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFIPLALIIICYLQVWLAIHT 0 0 VAQQQKESETTQKAERDVSRMVVVMIFAYIFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSEVSSSARTEVSSVSNSSVSPA* 0 >PIN_galGal Gallus gallus (chicken) Gt 0...2.2.0.0 indel x x x x 352 aa 000 nm no_ref genome pinopsin pineal non-visual 0 MSSNSSQAPPNGTPGPFDGPQWPYQAPQSTYVGVAVLMGTVVACASVVNGLVIVVSICYKKLRSPLNYILVNLAVADLLVTLCGSSVSLSNNINGFFVFGRRMCELEGFMVSLT 1 2 GIVGLWSLAILALERYVVVCRPLGDFQFQRRHAVSGCAFTWGWALLWSTPPLLGWSSYVPE 1 2 GLRTSCGPNWYTGGSNNNSYILSLFVTCFVLPLSLILFSYTNLLLTLRA 0 0 AAAQQKEADTTQRAEREVTRMVIVMVMAFLLCWLPYSTFALVVATHKGIIIQPVLASLPSYFSKTATVYNPIIYVFMNKQ 0 0 FQSCLLEMLCCGYQPQRTGKASPGTPGPHADVTAAGLRNKVMPAHPV* 0 >PIN_utaSta Uta stansburiana (lizard) Gt 0...2.2.0.0 indel x x x x 359 aa 000 nm 16543463 DQ100321 pinopsin pinopsin missing Anole genome 0 MVNEWSNATPGPFDGPQWPYLAPRSIYTSVAVLMGLVVVSAAFVNGLVIVVSIQYKKLRSPLNYILVNLAIADLLVTSFGSTLSFANNIYGFFVLGQTACEFEGFMVSLT 1 2 GIVGLWSLAILAFERYLVICKPVGDFRFQQRHAVFGCVFTWMWSLVWTLPPLFGWSSYVPE 1 2 GLRTSCGPNWYTGGSGNNSYIMALFVTCFALPLGMIIFSYASLLLTLRA 0 0 VATQQKEVETTQQAEKEVTRRVIAMVMAFLVCWLPYASFAMVVATNKDLVIQPALASLPSYFSKTATVYNPIIYVFMNKQ 0 0 FRSCLLSTMSCGHRPRGAQETTPAMISIPQGPTSALQGSRNKVTPSASEGSGNEAIPS* 0 >PIN_pheMad Phelsuma madagascariensis (gecko) Gt 0...2.2.0.0 indel x x x x 358 aa 000 nm no_ref AB022881 pinopsin 0 MHVQMANASQASLKNGTLSPFDGPQWPHRASRRVYTSLAALMGVVVLSASLANGLVIAVSVRFKRLRSPLNYILVNLATADLLVTFFGSIISFVNNAVGFFVFGKTACRFEGFMVSLT 1 2 GIVGLWSLAILAFERYLVICKPVGDFQFQRRHAVIGCLYTWGWSLIWTVPPLFGWSSYVPE 1 2 GLGTSCGPNWYMGGTNNNSYIVALFVTCFALPLSMILFSYANLLLTLRA 0 0 VAAQQKEQETTQRAEKEVTRMVITMVMAFLVCWLPYATFAMVVATTKDLSIQPGLASLPSYFSKTATVYNPIIYVFMNKQ 0 0 FRSCLLNTVSCGRIPQTMPGTPATTAVRGGFVLTSEGRGNKVASTELHS* 0 >PIN_podSic Podarcis sicula (lizard) Gt 0...2.2.0.0 indel x x x x 354 aa 000 nm 16688437 DQ013042 pinopsin pinopsin mRNA 0 MQASNASWVEVRNRTPGPFEGPQWPYLAPQSTYISVAVLMGLVVISATLVNGLVIVVSVQFKKLRSPLNYVLVNLAVADLLVTFFGSTISFVNNAQGFFIFGQATCEFEGFMVSLT 1 2 GIVGLWSLAILAFERYLVICKPVGDFRFPARHAVLGCAFTWGWSFVWTVPPLLGWSSYVPE 1 2 GLRTSCGPNWYSGGSSNNSYIMTLFVTCFAMPLSTILFSYANLLMTLRT 0 0 VAAQQKEQETTQRAEREVTRMVVAMVAAFLVCWLPYASFAMVVATHKDLAIRPALASLPSYFSKTATVYNPIIYVFMNKQ 0 0 FRSCLLYKMSCGHRALSSQDTTPAGISLPGRLTTSASKGSRNQVSPS* 0 >PIN_xenTro Xenopus tropicalis (frog) Gt 0...2.2.0.0 indel x x x x 346 aa 000 nm no_ref genome pinopsin 0 MRAGNMSAYEAPGPYDGPQWPHLAPRSTFLTVAAVMCMVVILAFFVNGLVIVVTLKYKKLRSPLNYILVNLAIANLLVTIFGSSVSFSNNVVGYFFMGKTMCEFEGFMVSLT 1 2 GIVGLWSLAILAFERYLVICKPMGDFRFQQKHAILGCSFTWVWSFIWTSPPLFGWCSYVPE 1 2 GLRTSCGPNWYTGGTNNNSYIMALFLTCFIMPLSTIIFSYSNLLMALRA 0 0 VAAQQKDSETTQRAEKEVTRMVIAMVLAFLICWLPYASFAVVVAVNKDVVIEPTVASLPSYFSKTATVYNPIIYVFMNKQ 0 0 FRNCLMTLLCCGRSFGDDETSSASGRTDVTSVSEAGGNKVTPA* 0 >PIN_bufJap Bufo japonicus (toad) Gt 0...2.2.0.0 indel x x x x 347 aa 000 nm 9537517 AF200433 pinopsin classifies oddly 0 MHSANMSALETPGPFEGPQWPHVAPRSTYLTVAVLMGMVVFLAFFVNGMVIVVSLKYKKLRSPLNYILVNLAVADILVTMFGSTVSFHNNIFGFFTLGKLVCELEGFVVSLT 1 2 GIVGLWSLAILAFERYIVICKPMGDFRFQQRHAVMGCAFTWIWAFLWTSPPLIGWCSYVPE 1 2 GLGTSCGPNWYTGGTNNNSYILALFTTCFMMPLTTIIFSYSNLLLALRA 0 0 VAAQQKESETTQRAEREVTRMVIAMVLAFLICWLPYAVFAIVMASNKNVVIDPTLASMPSYFSKTATVYNPVIYVFMNKQ 0 0 FRDCLTKLLCCGRNPFGEDETSTTSGRTDVTSVSEGGGNKVTPA* 0 >PIN_calMil Callorhinchus milii (elephantfish) Gt 0...2.2.0.0 indel x x x x 093 aa 000 nm no_ref genome fragment no petMar 0 FGSTVSFSNNINGYFVLGETVCQFEGFMVSLT 1 2 GIVGLWSLAILAFERYIVICKPMGDFRFQQKHAVWGCLFTWLWSLFWTLPPLFGWCSYVPE 1 >VAOP_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel +INPP5A -NXK6 +C10orf61 +ALDH18A1 393 aa 000 nm no_ref genome TCTN3 exon 1 genbank error 0 MDVFRALGNESLLSNSSGPARWDPFHHPLDSIQPWHFRLVAAVMFVVTSLSLAENLAVILVTFKFKQLRQPVNYVIVNLSVADFLVSLTGGTISFLANLKGYFYMGHWACVLEGFAVTFF 1 2 GIVALWSLALLAFERYIVICRPVGNMRLRGKHAAQGIAFVWTFSFIWTIPPTMGWSSYTTSKIGTTCEPNW 2 1 YSGAYNDRSYIIAFFTTCFIVPLLVILVSYGKLLQKLRK 0 0 VSNTQGRLRTARKPERQVTRMVVVMIIAFLICWMPYAVFSILATAYPSIELDPHLAAIPAFFSKTATVYNPIIYVFMNKQ 0 0 FRMCLIQMFKCSAIETAESNMNPTSERATLTQDKRDSQLSVMAVRSTILKRKTGDEHRADDLWLFRQLQKPKCVPCRAGDGS* 0 >VAOP_anoCar Anolis carolinensis (lizard) Gt 0...2.1.0.0 indel +INPP5A -NXK6 +GPR125 +KNDC1 389 aa 000 nm no_ref genome vertebrate ancient 0 MAGLRREAENDSWLFDPSSSSAPFDPFLQPLDIIEPWNFHLISALMFVVTLFSLSENFTVILVTIKFKQLRQPLNYVIVNLSVADFLVSLIGGTISFSTNLKGYFYMGHWACVLEGFAVTFF 1 2 GIVALWSLALLAFERYVVICRPLGNMRLNGKHAALGVAFVWIFSFIWTVPPTMGWSSYTTSKIGTTCEPNW 2 1 YSGDYNDHTFIITFFTTCFILPLLVILVSYGKLMRKLRK 0 0 VSDTQGRLGTTRKPERQVTGMVVIMILAFLICWSPYAAFSILVTACPSIELDPRLAAIPAFFSKTATVYNPVIYVFMNNQ 0 0 FRKCLVQLFQCSSQETMDANVNPISEKDTLTHTKHCGEMSTVAAHVIVFNPRSEDEQGSCQSFAQLAISENKVYPL* 0 >VAOP_xenTro Xenopus tropicalis (frog) Gt 0...2.1.0.0 indel - +GSTO2 -C10orf92 - 383 aa 000 nm no_ref genome vertebrate ancient new 0 MPTNVSLLATPENSTVWNPFTGPLKTIEAWNFHLLAALMFVVTSLSIAENFIVILVTAKFKQLRQPLNYIIVNLSVADFLVSVIGGTISIATNSRGYFYLGSWACVLEGFAVTFF 1 2 GIVALWSLSVLAFERYIVICRPLGNLRLQGKHSALAIIFVWVFSFVWTIPPTMGWSSYTTSKIGTTCEPNW 2 1 YSGEMRDHTYIITFLTTCFVFPLLVIFMSYGKLMRKLRK 0 0 VSDTQGRLGSTRKPEKEVTRMVVIMILAFLICWTPYAAFSILITAHPTIDLDPRLAAIPAFFAKTASMYNPIIYVYMNKQ 0 0 FRRCLYQMFNINDPEAKESNLNPTSERGVLTRNNNGGEMLAIATHITSSAVTNREEEKSSSNSFAHIPVSDNKVCPM* >VAOP_danRer Danio rerio (zebrafish) Gt 0...2.1.0.0 indel - - - - 378 aa 000 nm 17067577 NM_131586 vertebrate ancient valop vertebrate assembly missing exon 3 0 MEASSAAVNAVSPAEDPFSAPLSSIAPWNYSVLAALMFVVTALSLSENFTVMLVTFRFQQLRQPLNYIIVNLSLADFLVSLTGGSISFLTNYHGYFFLGKWACVLEGFAVTFF 1 2 GIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLVFVWSFSFIWTVPPVLGWSSYTVSRIGTTCEPNW 2 1 YSGNFHDHTFIITLFSTCFIFPLGVIIVCYCKLIRKLRK 0 0 VSNTHGRLGNARKPERQVTRMVVVMIVAFMVAWTPYAAFSIIITAHPSMHVDPRLAAIPAFVAKTAAVYNPIIYVFMNKQ 0 0 FRKCLVQLLSCSKVTVVEGNNNQTTERAGMTSGSNTGEMSAIAARVSVPKTEENPGDRSTFSHIPIPENKVCPM* >VAOP_takRub Takifugu rubripes (teleost) Gt 0...2.1.0.0 indel +INPP5A -NXK6 - +KNDC1 362 aa 000 nm no_ref genome vertebrate ancient 0 MESLSLSVNGVSYTVAAELAPTNDPFTGPINNIAQWNFTILAVLMFVVTSLSLCENFLVMFITFKFKQLRQPLNYIIVNLAIADFLVSLTGGLISFLTNARGYFFLGRWACVLEGFAVTYF 1 2 GIVAMWSLAVLSFERFFVICRPLGNMRLQAKHAAIGLLFVWTFSFVWTFPPVLGWNRYTVSKIGTTCEPDW 2 1 YSNNMTSHSYIITFFSTCFILPLGIIFFCYGKLLRKLRK 0 0 VSHGRLATARKPERQVTRMVVVMIVAFMVAWTPYATFAILVTIHPTIELDPRLASIPAFFSKTAAVYNPIIYVFMNKQ 0 0 FRKCLIQHFIGMGVMAESNMNPTSERPGITAESQTGEMSAIAARVPVGATAALHSDGSPTDCGSLAQLPIPENKVCPI* 0 >VAOP_rutRut Rutilus rutilus (minnow) Gt 0...2.1.0.0 indel x x x x 383 aa 000 nm 12906786 AY116411 vertebrate ancient vertebrate 0 MELFPVAVNGVSHAEDPFSGPLTFIAPWNYKVLATLMFVVTAASLSENFAVMLVTFRFTQLRKPLNYIIVNLSLADFLVSLTGGTISFLTNYHGYFFLGKWACVLEGFAVTYF 1 2 GIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLLFVWTFSFIWTIPPVLGWSSYTVSKIGTTCEPNW 2 1 YSGNFHDHTFIIAFFITCFILPLGVIVVCYCKLIKKLRK 0 0 VSNTHGRLGNARKPERQVTRMVVVMIVAFMVAWTPYAAFSIVVTAHPSIHLDPRLAAAPAFFSKTAAVYNPVIYVFMNKQ 0 0 FRKCLVQLLRCRDVTIIEGNINQTSERQGMTNESHTGEMSTIASRIPKDGSIPEKTQEHPGERRSLAHIPIPENKVCPM* 0 >VAOP_calMil Callorhinchus milii (elephantfish) Gt 0...2.1.0.0 indel x x x x 080 aa 000 nm no_ref genome fragment 0 VASTQGRLGVARKPEKQVTRMVIVMILAFLFCWTPYAAFSITVTACPTIKLDPRLAAIPAFFSKTATVYNPIIYVFMNKQ 0 >VAOP_petMar Petromyzon marinus (lamprey) Gt 0...2.1.0.0 indel x x x x 445 aa 000 nm 9427550 U90667 vertebrate ancient exons 123 in traces pineal gland-specific 0 MDALQESPPSHHSLPSALPSATGGNGTVATMHNPFERPLEGIAPWNFTMLAALMGTITALSLGENFAVIVVTARFRQLRQPLNYVLVNLAAADLLVSAIGGSVSFFTNIKGYFFLGVHACVLEGFAVTYF 1 2 GVVALWSLALLAFERYFVICRPLGNFRLQSKHAVLGLAVVWVFSLACTLPPVLGWSSYRPSMIGTTCEPNW 2 1 YSGELHDHTFILMFFSTCFIFPLAVIFFSYGKLIQKLKK 0 0 ASETQRGLESTRRAEQQVTRMVVVMILAFLVCWMPYATFSIVVTACPTIHLDPLLAAVPAFFSKTATVYNPVIYIFMNKQ 0 0 FRDCFVQVLPCKGLKKVSATQTAGAQDTEHTASVNTQSPGNRHNIALAAGSLRFTGAVAPSPATGVVEPTMSAAGSMGAPPNKSTAPCQQQGQQQQQQGTPIPAITHVQPLLTHSESVSKICPV* 0 >PPIN_anoCar Anolis carolinensis (lizard) Gt 0...2...0.0 indel -CPEB2 -CACNA2D3 +SELK +ACTR8 346 aa 000 nm no_ref genome parapinopsin syntenic deleted in chicken 0 MDSLDTNTLSPNASTVRVVLMPRIGYTIIAIIMATSCTLSVILNTAVIAITIKYRQLRQPINYSLVNLAIADLGAALLGGSLNVETNAVGYYNLGRVGCVTEGFAMAFF 1 2 GIVALCTIAVIAVDRAIVIAKPMGTITFTTRKAMIGVAVSWIWSLVWNTPPLFGWGGYQMEGVMTSCAPDWANSDPINVSYIICYFLFCFTIPFITILASYGYLIWTLRQ 0 0 VAKVGLAQRGSTTKAEAQVSRMVIVMVMAFLICWLPYATFALVVVGNPQIYINPIIATIPMYMAKSSTFYNPIIYIFMNKQ 0 0 FRDCLVRCLLCGRNPCASEQTDEDDLEVSTIAPAPSSRRGKVAPV* 0 >PPIN_xenTro Xenopus tropicalis (frog) Gt 0...2...0.0 indel - - +SELK - 349 aa 000 nm no_ref genome parapinopsin bistable UV lamprey pineal broken contigs 0 MADEALLPPMMNVTNEEMHPGKVLMPRIGYTILALIMAVFCAAALFLNVTVIVVTFKYRQLRHPINYSLVNLAIADLGVTVLGGALTVETNAVGYFNLGRVGCVIEGFAVAFF 1 2 GIAALCTIAVIALDRVFVVCKPMGTLTFTPKQALAGIAASWIWSLIWNTPPLFGWGSYELEGVMTSCAPNWYSADPVNMSYIVCYFSFCFAIPFLIIVGSYGYLMWTLRQ 0 0 VAKLGVAEGGTTSKAEVQVSRMVIVMILAFLVCWLPYAAFAMTVVANPGMHIDPIIATVPMYLTKTSTVYNPIIYIFMNKQ 0 0 FQECVIPFLFCGRNPWAAEKSSSMETSISVTSGTPTKRGQVAPA* 0 >PPIN_ictPun Ictalurus punctatus (catfish) Gt 0...2...0.0 indel x x x x 347 aa 000 nm no_ref genome parapinopsin parapinopsin index sequence 0 MASIILINFSETDTLHLGSVNDHIMPRIGYTILSIIMALSSTFGIILNMVVIIVTVRYKQLRQPLNYALVNLAVADLGCPVFGGLLTAVTNAMGYFSLGRVGCVLEGFAVAFF 1 2 GIAGLCSVAVIAVDRYMVVCRPLGAVMFQTKHALAGVVFSWVWSFIWNTPPLFGWGSYQLEGVMTSCAPNWYRRDPVNVSYILCYFMLCFALPFATIIFSYMHLLHTLWQ 0 0 VAKLQVADSGSTAKVEVQVARMVVIMVMAFLLTWLPYAAFALTVIIDSNIYINPVIGTIPAYLAKSSTVFNPIIYIFMNRQ 0 0 FRDYALPCLLCGKNPWAAKEGRDSDTNTLTTTVSKNTSVSPL* 0 >PPIN_danRer Danio rerio (zebrafish) Gt 0...2...0.0 indel - - +SELK - 338 aa 000 nm no_ref XM_681591 parapinopsin parapinopsin 0 MESETSTAASGSIAEVMPRMGYTILAVIIGVFSVCGVILNVTVITVTLKYKQLRQPLNFALVNLAVADLGCAVFGGLPTVVTNAMGYFSLGRVGCVLEGFAVAFF 1 2 GIAALCSVAVIALERCMVVCRPVGSISFQTRHAVFGVAVSWLWSFIWNTPPLFGWGRLQLEGVRTSCAPDWYSRDLANVSFIVCYFLLCFALPFSVIVYSYTRLLWTLRQ 0 0 VSRLQVCEGGSAARAEAQVSCMVVVMILAFLLTWLPYASFALCVILIPELYIDPVIATVPMYLTKSSTVFNPIIYIFMNRQ 0 0 FRDRALPFLLCGRNPWAAEAEEEEEETTVSSVSRSTSVSPA* 0 >PPIN_oncMyk Oncorhynchus mykiss (trout) Gt 0...2...0.0 indel x x x x 347 aa 000 nm no_ref genome parapinopsin 0 MDHQQLLPNLHGNISSSPGSVSEALLSRTGFTILAVIIGVFSVSGVCMNVLVIMVTMRHRKLRQPLNYALVNLAVADLGCALFGGLPTMVTNAMGYFSMGRLGCVLEGFAVAFF 1 2 GIAGLCSVAVIAVDRYVVVCRPMGAVMFQTRHAVGGVVLSWVWSFLWNTPPLFGWGSFELEGVRTSCSPNWYSREPGNMSYIILYFLLCFAIPFSIIMVSYARILFTLHQ 0 0 VSKLKVLEGNSTTRVEIQVVRMVVVMVMAFLLSWLPYAAFALSVILDPSLHINPLIATVPMYLAKSSTVYNPIIYVFMNRQ 0 0 FRDCAVPFLLCGLNPWASEPVGSEADTALSSVSKNPRVSPQ* >PPIN_calMil Callorhinchus milii (elephantfish) Gt 0...2...0.0 indel x x x x 109 aa 000 nm no_ref genome fragment 0 MDPHNRSANLSEGPGLGGGGAVPGWGPSVRAPLSLVMAVISLSSIVLNSLAIAVVLRFQVLQQPLNYALLSLASADLGTAATGGVLSTVCTALGSFVLGRHSCVAEGFF 1 >PPIN_petMar Petromyzon maritimus (lamprey) Gt 0...2...0.0 indel x x x x 344 aa 000 nm no_ref genome parapinopsin bistable pineal UV/green 0 MENLTSLDLLPNGEVPLMPRYGFTILAVIMAVFTLASLVLNSTVIIVTLRHRQLRHPLNFSLVNLAVADLGVTVFGASLVVETNAVGYFNLGRVGCVIEGFAVAFF 1 2 GIAALCTIAVIAVDRFVVVCKPLGTLMFTRRHALLGITWAWLWSFVWNTPPLFGWGSYKLEGVRTSCAPDWYSRDPANVSYIVSFFSFCFAIPFLVIVVAYGRLLWTLHQ 0 0 VAKLGMGESGSTAKAEAQVSRMVVVMVVAFLVCWLPYALFAMIVVAKPGVYIDPVIATLPMYLTKTSTVYNPIIYIFMNRQ 0 0 FRDCAVPFLLCGRNPWAEPSSESATTASTSATSVTLASVPGQVSPS* 0 >PPIN_letJap Lethenteron japonicum lamprey Gt 0...2...0.0 indel x x x x 344 aa 000 nm 14981504 AB116380 parapinopsin bistable pineal UV/green 0 MENLTSLDLLPNGEVPLMPRYGFTILAVIMAVFTIASLVLNSTVVIVTLRHRQLRHPLNFSLVNLAVADLGVTVFGASLVVETNAVGYFNLGRVGCVIEGFAVAFF 1 2 GIAALCTIAVIAVDRFVVVCKPLGTLMFTRRHALLGIAWAWLWSFVWNTPPLFGWGSYELEGVRTSCAPDWYSRDPANVSYITSYFAFCFAIPFLVIVVAYGRLMWTLHQ 0 0 VAKLGMGESGSTAKAEAQVSRMVVVMVVAFLVCWLPYALFAMIVVTKPDVYIDPVIATLPMYLTKTSTVYNPIIYIFMNRQ 0 0 FRDCAVPFLLCGRNPWAEPSSESATAASTSATSVTLASAPGQVSPS* 0 >PPINa_cioInt Ciona intestinalis (tunicate) Gt 0...2...0.0 indel -HOXB1 +HHEX +CUL4A - 391 aa 000 nm 11591373 NM_001032555 parapinopsin Ci-opsin odd exons larval ocellus 0 MDHDVTPTVDLTDGVPQCKDLNPYVLKGDGWVPQHISRANRSTYSFLCVYMTFVFLLSCSLNILVIVATLKNK 0 0 VLRQPLNYIIVNLAVVDLLSGFVGGFISIAANGAGYFFWGKTMCQIEGYFVSNFGVTGLL 0 0 SIAVMAFERYFVICKPFGPVRFEEKHSIFGIV 0 0 ITWVWSMFWNTPPLIFWDGYDTEGLGTSCAPNWFVKEKRERLFIILYFVFCFVIPLAVIMICYGKLILTLRQ 0 0 IAKESSLSGGTSPEGEVTKMVVVMVTAFVFCWLPYAAFAMYNVVNPEAQ 0 0 IDYALGAAPAFFAKTATIYNPLIYIGLNRQ 0 0 FRDCVVRMIFNGRNPWVDELVGSQVSSTGSQLTAVSSNKVAPA* 0 >PPINb_cioInt Ciona intestinalis (tunicate) Gt 0...2...0.0 indel -TMEM165 +FUT4 - - 353 aa 000 nm no_ref genome parapinopsin jgi gene model wrong both ends 0 MTTAETTTECYEKNPYIRNEMGWVPKHILIAERHIYTILAVYMTFIFLLAVSLNGFVIIATMKNK 0 0 KLRQPLNYIIINLSIADFLSGLVGGFIGMISNSAGYFYFGKTVCILEGYIVSVA 1 2 GVCGLMSISVMAFERYFVVCKPYGPFTLTNTHAAL 1 2 GIGFTWTWSVLWSTPGLIWLDGYVPEGLGTSCAPNWFSKNK 2 1 SERIFIFVYFVFCFFIPLLVIIICYGKIVLFLKQVSLY 0 0 ATRQSSASSNRQADNKVTKMVLVMISAFLICWTPYGVLSLYNAINPDKQ 0 0 LDYGLGAVPVFFAKTANIYNPLIYIGLNKQ 0 0 FRDGVIKMVFRGRNPWAEEMSTQQRQRSTEAGQPIVSNEV* 0 >PARIE_utaSta Uta stansburiana (lizard) Gd+Go 0...2...0.0 indel x x x x 347 aa 522 nm 16543463 DQ100320 parietopsin shift in counterion Gt + Go 0 MENDSSLATELAEGAIVKPTIFPKAGYGVLAFLMFLNALFSIFNNSLVIAVTLKNPQLRNPINIFILNLSFSDLMMSLCGTTIVIATNYYGYFYLGRKFCIFQGFAVNYF 1 2 GIVSLWSLTILAYERYNVVCQPLGTLQMSTKRGYQLLGFIWVFCLFWAVVPLFGWSSYGPEGVQTSCSIGWEERSWSNYSYLIVYFLSCFFIPVLIIGFSYGNVIRSLHG 0 0 LNKKVEQLGGKSSPEEEFRAVIMVLVMVVAFLICWLPYTVFALIVVFNPALNISPLAATIPTYLSKTSPVYNPIIYIFLNKQ 0 0 FRDCAVEFITCGQVVLTSPEEDISTSAIPVEGKGPCKINQVTPV* 0 >PARIE_anoCar Anolis carolinensis (lizard) Gd+Go 0...2...0.0 indel +EEA1 -FLJ46688 +BTG1 - 347 aa 000 nm no_ref genome parietopsin Go like scallop, gusducin not transducin 0 MENESSLVLEGAEGYIVRPTIFPRAGYGVLAFLMFINALFSLFNNFLVIAVTLKNPQLRNPINIFILNLSFSDLMMSICGTTIVIATNYHGYFYLGRRFCIFQGFAVNYF 1 2 GIVSLWSLTILAYERYNVVCQPLGTLQMSTQRAYQLLGFIWVFCLFWAVVPLFGWSSYGPEGVQTSCSIGWEERSWNNYSYLIVYFLSCFFIPVLIIGFSYGNVIRSLHG 0 0 LNKKVEQLGGKSNPEEEFRAVIMVLVMVVAFLICWLPYTLFALTVVFNPALNISPLAATIPTYLSKTSPVYNPIIYIFLNKE 0 0 FRECAVEFITCGKVVLTSPEEDISTSAISDEGIAPCKINQVTPV* 0 >PARIE_xenTro Xenopus tropicalis (frog) Gd+Go 0...2...0.0 indel -lum -DCN - - 346 aa 000 nm 16543463 NM_001045791 parietopsin 0 MDGNSTTPGIAVNLTVMPTIFPRSGYSILSFLMFLNAVFSICNNAIVILVTLKHPQLRNPINIFILNLSFSDLMMALCGTTIVVSTNYHGYFYLGKQFCIFQGFAVNYF 1 2 GIVSLWSLTLLAYERYNVVCEPIGALKLSTKRGYQGLVFIWLFCLFWAIAPLFGWSSYGPEGVQTSCSIGWEERSWSNYSYIISYFLTCFIIPVGIIGFSYGSILRSLHQ 0 0 LNRKIEQQGGKTNPREEKRVVIMVLFMVLAFLICWLPYTVFALIVVINPQLYISPLAATLPTYFAKTSPVYNPIIYIFLNKQ 0 0 FRTYAVQCLTCGHINLDSLEEDTESVSAQAENMLTPKTNQVAPA* 0 >PARIE_takRub Takifugu rubripes (teleost) Gd+Go 0...2...0.0 indel -HSP90B1 +NT5DC2 -KCND3 -FLNC 351 aa 000 nm 16543463 genome parietopsin 0 MDSNSTPWSSPPAPLQAEAVTVAPTIFPRVGYSILSFLMFINTVLSVFNNSLAIAVMLKNPSLLQPINIFILSLAVSDLMIGLCGSLVVTITNYHGSFFIGHTACVFQGFAVNYF 1 2 GLVSLCTLTLLAYERYNVVCKPRAGLKLTMRRSIIGLLFVWTFCLFWAVTPLLGWSSYGPEGVQTSCSLAWEERSWNNYSYLILYTLLCFIFPVGVIIYCYCKVLTSMNK 0 0 LNKSVELQGGLSCRRENKHAINMVLAMIIAFFVCWLPYTALSVVVVVDPELHIPPLVATMPMYFAKTSPVYNPIIYFLSNKQ 0 0 FRDATLEVLSCSRYIPHASSRVSINMRSLNRRSVNTHSKVSPL* 0 >PARIE_gasAcu Gasterosteus aculeatus (stickleback) Gd+Go 0...2...0.0 indel -HSP90B1 +NT5DC2 -KCND3 -FLNC 361 aa 000 nm no_ref genome parietopsin 0 MDSNSTLWSSGSPPPSIHGKMLTITPTIFPRVGYSILSFLMFINTVLTVFNNVLVITVLVRNPSLLQPMNVFILSLAVSDLMIGLCGSLVVTITNYHGSFFIGHTACIFQGFAVNYF 1 2 GLVSLCTLTLLSYERYNVVCRPRNALKLSMRRSIHGLLIVWTFCLFWAVAPLFGWSGYGPEGVQTSCSLAWEERSWSNYSYLVLYTLLCFIVPVAVIIYCYAKVLTSMNT 0 0 LNRSVEVQGGRSSQKENDHAVSMVLAMIIAFFSCWLPYTALSVVVVVDPTLYIPPLVATMPMYFAKTSPVYNPIIYFLSNKQ 0 0 FRDAALEMLSCGRYIAHMPNTVSINMRSLNRRSRLSSLSRNVNSHSKVLPL* 0 >PARIE_danRer Danio rerio (zebrafish) Gd+Go 0...2...0.0 indel - +NT5DC2 +FBXL13 - 337 aa 000 nm 16543463 genome parietopsin 0 MENFAKTELTMMVQPTIFPRVGYSILSYLMFINTTLSVFNNVLVIAVMVKNLHFLNAMTVIIFSLAVSDLLIATCGSAIVTVTNYEGSFFLGDAFCVFQGFAVNYF 1 2 GLVSLCTLTLLAYERYNVVCKPMAGFKLNVGRSCQGLLLVWLYCLFWAVAPLLGWSSYGPEGVQTSCSLGWEERSWRNYSYLILYTLMCFILPTVIITYCYSNVLLTMRK 0 0 INKSIECQGGKNCAEDNEHAVRMVLAMIIAFFICWLPYTAISVLVVVNPEISIPPLIATMPMYFAKTSPVYNPIIYFLTNKR 0 0 FRESSLEVLSCGRYISRETGGPLMGSSMQRGQSRVNPV* 0 >PARIE_petMar Petromyzon marinus (lamprey) Gd+Go 0...2...0.0 indel x x x x 082 aa 000 nm no_ref genome fragment 0 LNKKIKRVGGHPDPREEMRATVMVLAMVGAFLACWLPYTVLALCVVLAPGTQIPPLVATLPMYFAKTSPMYNPIIYFFLNPQ 0 >ENCEPH_homSap Homo sapiens (human) Gt 0...2...0.0 indel -EXO1 -WDR64 -KMO +FH 403 aa 000 nm 12242008 NM_014322 parietopsin OPN3 with intron loss 0 MYSGNRSGGHGYWDGGGAAGAEGPAPAGTLSPAPLFSPGTYERLALLLGSIGLLGVGNNLLVLVLYYKFQRLRTPTHLLLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSGSLF 1 2 GIVSIATLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPLGVIAHCYGHILYSIRM 0 0 LRCVEDLQTIQVIKILKYEKKLAKMCFLMIFTFLVCWMPYIVICFLVVNGHGHLVTPTISIVSYLFAKSNTVYNPVIYVFMIRK 0 0 FRRSLLQLLCLRLLRCQRPAKDLPAAGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDKTNGSKVDVIQVRPL* 0 >ENCEPH_monDom Monodelphis domestica (opossum) Gt 0...2...0.0 indel -EXO1 -WDR64 -KMO +FH 411 aa 000 nm no_ref genome encephalopsin OPN3 extra intron alt splicing 0 MYSDNSSDDGGGGYWGSGRAGGASGTGVTGEPGPEGSPRQAPLFSPGTYELLALLIATIGLLGLCNNLLVLVLYYKFQRLRTPTHLFLVNISFNDLLVSLFGVTFTFVSCLRSGWVWDSVGCAWDGFSNTLF 1 2 GIVSIMTLTVLAYERYNRIVHAKVINFSWAWRAITYIWLYSLVWTGAPLLGWNRYTLEIHGLGCSVDWKSKDPNDSSFVIFLFFGCLMLPVGVMAYCYGHILYAIRM 0 LRCVEELQTIQVIKILRYEKKVAKMCFLMIAIFLFCWMPYAVICLLVANGYGSLVTPTVAIIASLFAKSSTAYNPIIYIFMSRK 0 0 FRRCLLQLLCFRLLKFQQPKKDRPVIRTEKQIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDETQMIDENDKNSGTKVNVIQVRPL* 0 >ENCEPH_galGal Gallus gallus (chicken) Gt 0...2...0.0 indel -EXO1 -WDR64 -PIGM +RGS7 396 aa 000 nm no_ref genome encephalopsin OPN3 0 MHSGNGTGATSRPQLAAAGHEVPGERPLFSAGTYELLALLIATIGTLGVCNNLLVLVLYYKFKRLRTPTNLFLVNISLSDLLVSVCGVSLTFMSCLRSRWVWDAAGCVWDGFSNSLF 1 2 GIVSIMTLTVLAYERYIRVVHAKVIDFSWSWRAITYIWLYSLAWTGAPLLGWNRYTLEIHGLGCSMDWKSKDPNDTSFVLLFFLGCLVAPVVIMAYCYGHILYAVRM 0 0 LRCVEDFQTSQVIKLLKYEKKVAKMCFLMISTFLICWMPYAVVSLLVTYGYSNLVTPTVAIIPSFFAKSSTAYNPVIYIFMSRK 0 0 FRQCLLQLLCFRLMRFQRIMKEPSGAGNVKPIRPIVMSQKVGDRPKKKVTFSSSSIIFIIASDDTQQIDDNSKHNGTKVNVIQVKPL* 0 >ENCEPH_anoCar Anolis carolinensis (lizard) Gt 0...2...0.0 indel -EXO1 -WDR64 -PIGM +RGS7 408 aa 000 nm no_ref genome encephalopsin OPN3 0 MFSANGTRSGAGSDLEPGPGQQQQQREASEEEERGAGLSPFSAGTYELLALLVAAIGLLGLCNNLLVLVLYAKFKRLRTPTHLFLVNISLSDLLVSLFGVSFTFGSCLRHRWVWDAAGCVWDGFSNSLF 1 2 GIVSIMTLTVLAYERYIRVVHARVIDFSWSWRAITYIWLYSLAWTGAPLLGWNHYTLEIHGLGCSVDWQSKEPSDSSFVLFFFLGCLAAPVGIMAYCYGHILHAIRM 0 0 LRCVEDLQSIQVIKILRYEKKVAKMCFLMVTTFLICWMPYAVVSLLIAYGYGHLITPTVAIIPSFFAKSSTAYNPVIYIFMSRK 0 0 FRRCLVQLFCVQFLRFKRTLKEQPAIESNKPIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDDTEQIDVSTKCSDTKINVIQVKPL* 0 >ENCEPH_takRub Takifugu rubripes (teleost) Gt 0...2...0.0 indel -ABLIM1 +PTK7 -KMO +IDE 388 aa 000 nm no_ref genome encephalopsin TMT multiple tissue circadian clock 0 MPVTNGSHNNSISWLHSKDMFTEDTYHFLALIVATVGFLGLVNNLLVLILYCKFKRLQTPTNLLFFNTSLCHFVFSLLAITFTFMSCVRGSWAFSVEMCVFHGFSKNLL 1 2 GIVSFGTLTVVAYERYARVVYGKYVNSSWSKRSITFVWVYSLAWTGFPLIGWNLYTFETHKLDCSFEWTATDPKDTAFVLLFFLACITLPLSIMAYCYGYILYEIQK 0 0 LRSVKNIQNFQEITILDYEIKMAKMCLLMMLTFLIGWMPYTILSLLVTSGYSKFITPTITVMPSLLAIASAAYNPVIHIFTIKK 0 0 FRQCLVQLLPPINFHPPINPPINNFWRLLKNLNGRLAMKKVKPVLGKGRSHNRPEKKVPPINFSSSDFFTRTTSDTGTHGITESTKGKRTNVRLIQVHPL* 0 >ENCEPH4a_takRub Takifugu rubripes (teleost) Gt 0...2...0.0 indel -CALD1 +TNK2 -RAB18 +ABI1 403 aa 000 nm 12670711 AF402774 encephalopsin TMT multiple tissue circadian clock 0 MIVSNVSLSGCAGVNGAVCAAEGHQAGGSDRSTLTPTGNLVVSVFLGFIGTFGLVNNLLVLVLFCRYKMLRSPINLLLMNISISDLLVCVLGTPFSFAASTQGRWLIGEAGCVWYGFANSLF 1 2 GVVSLISLAVLSFERYSTMMTPTEADPSNYCKVCLGITLSWVYSLVWTVPPLFGWSSYGPEGPGTTCSVNWTAKTTNSISYIICLFVFCLIVPFLVIVFCYGKLLCAIRQ 0 0 VSGINASTSRKREQRVLCMVVIMVICYLLCWLPYGVVALLATFGPPDLVTPEASIIPSVLAKSSTVINPIIYVFMNKQ 0 0 FYRCFLALLCCQDPRSGSSMKSSSKVATKAKGVTPTGQRRTDFLYMVASLGRPAATIPQLGPSFDATNDFTKPPSSDTIKPVVVSLAAHCDG* >ENCEPH4b_takRub Takifugu rubripes (teleost) Gt 0...2...0.0 indel +TFRC +CHES1 -MYEOV2 -ARHGAP21 407 aa 000 nm no_ref genome encephalopsin 0 MIVCNVSLSCAHCPGEGTAANDAYAQASGSLATPTLSQRGHLVVAVCLGFIGTVGFLSNFLVLALFCRYRALRTPMNLMLVSISASDLLVSVLGTPFSFAASTQGRWLIGRAGCVWYGFVNACL 1 2 GIVSLISLAVLSYERYCTMVSSTIASNRDYRPVLGGICFSWFYSLAWTVPPLLGWSRYGPEGPGTTCSVDWRTQTPNNISYIVCLFTFCLLLPFFVILYSYGKLLHTIRQ 0 0 VRRVSSTVTRRREHRVLVMVVAMVVCYLICWLPYGVTALLATFGPPNLLTPEATITPSLLAKFSTVINPFIYIFMNKQ 0 0 FYRCFRAFLNCSTPKRDSTVRTFTRISLRALRQDQQQKGSALAPSSARPTPNSIHESSLKGSHSTPSNGGAAAAKSPAANRSKPKLILVAHYRE* 0 >ENCEPH_gasAcu Gasterosteus aculeatus (stickleback) Gt 0...2...0.0 indel -LDOC1L +CDC42EP3 -KMO +IDE 389 aa 000 nm no_ref genome encephalopsin OPN3 0 MNPDNGTREERSTDHSIFAVGTYKLLAFAIGTIGVFGFCNNVVVIVLYCKFKRLRTPTNLLVVNISLSDLLVSVIGINFTFVSCIRGGWTWSRATCIWDGFSNSLF 1 2 GIVSIMTLASLAYERYIRVVHAQVVDFPWAWRAIGHIWLYSLVWTGAPLLGWNRYTLEIHRLGCSLDWASKDPNDASFILLFLLACFFVPVGIMIYCYGNILYAVQM 0 0 LRSIQDLQTVQIIKILRYEKKVAVMFLLMISCFLLCWTPYAVVSMMEAFGRKNMVSPTVAIIPSFFAKSSTAYNPLICVFMSRK 0 0 FRRCLMQLLCSRVTCLQCNLKERPLAPVQRPIRPIVVSAACGGGRVRPKKRVTFSSSSIVFIITRNDIRHTDVTSNTRESSEANVFQVRPL* 0 >ENCEPH_calMil Callorhinchus milii (elephantfish) Gt 0...2...0.0 indel x x x x 097 aa 000 nm no_ref genome fragment 0 MNPTNSTEPQEEHLFSPNTYKLLAVIIGTIGIVGFCNNILVLLLYYKFKRLRTPTNLLLVNISVSDLLVSVFGLSFTFVSCTQGRWGWDSAACVWDG >ENCEPH4_calMil Callorhinchus milii (elephantfish) Gt 0...2...0.0 indel x x x x 177 aa 000 nm no_ref genome fragment 0 MLNSSPNSSPSLPLSQVGWTGLSRTGLTVVAVCLGIIMVLGFLNNLLVLVLFCKYKVLRSPMNMLLLNISVSDMLVCICGTPFSFAASVQGRWLVGEQGCKWYGFANSLF 1 0 REHRILLMVISMVTFYLLCWLPYGTVALIGTFGNADLITPTCSVIPSILAKSSTVINPVIYVIMNKQ 0 >ENCEPH5_calMil Callorhinchus milii (elephantfish) Gt 0...2...0.0 indel x x x x 070 aa 000 nm no_ref genome fragment AQTREHRILLMVISMVTFYLLCWLPYGTVALIGTFGNADLITPTCSVIPSILAKSSTVINPVIYVIMNKQ 0 >ENCEPH_squAca Squalus acanthias (dogfish) Gt 0...2...0.0 indel x x x x 202 aa 000 nm no_ref genome fragment 0 MNAANSTDTREESLFSPGTYQVLAVIIGTIGVVGFCNNLLMLVLYCKFKRLRTPTNLFLVNISISDLLLSVFGVIFTFVSCVKGRWVWDSAACVWDGFSNCLF 1 2 GISSIMSLTVLAYERYIRVVNATAIDFSWAWRAITYIWLYSLAWTGAPLIGWNSYTLELHRLGCSVNWDSRNPSDTSFVLFLFLGCLLCPIGVIAYCYG >ENCEPH_petMar Petromyzon marinus (lamprey) Gt 0...2...0.0 indel x x x x 293 aa 000 nm no_ref genome fragment 0 MQSPKQDSLHYAGDTGAKAAPDSAQGNASALGSNFLLHGGDLGEGSTAFSAATFRLLAGVVGTIGVAGFLNNLLLVALFVGFKRLQTPTNLLLVNISLSDLLVSVFGNTLTLVSCVRRRWVWGNGGCVWDGFSNSLF 1 2 GIVSISTLTALSYERYARLIKAQVLDFSWAWRAVTYTWLYSAAWTGAPLLGWSRYVLEKHGLGCSIDWASSNPPDAAFVLFFFLGCLAAPLLVMGFCFGRIALAITQ 0 0 CWSPYAVASLFVASGFEHLVSPPVSIVPSLLAKSNAVCNPLLFLLMSGN 0 >ENCEPH4_braFlo Branchiostoma floridae (amphioxus) Gt 0...2...0.0 indel -ZFYVE1 +RTF1 -CES1 -POMT2 402 aa 000 nm 12435605 AB050608 encephalopsin Amphiop4 new exon 12 and 34 + perfect fit 0 MALYNNTSSPSQDLLWDAPYSQGHIWDNSSASNSSEDVMDQGKVELQDFSDAGYTAIATCLALI 1 2 GFVGFTNNFVVILLIGCHRQLRTPFNLLLLNMSVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANSLF 1 2 GIVSLVTLSALAFERYCVVVRSSDMLTYKSSLVVITFIWLYSLLWTSLPLLGWSSYQFEGHN 0 0 VGCSVNWVQHNPDNVSYIVTLMVTCFFVPMVVVCWSYAWIWRTVRM 0 0 SSEAKPECGNSQNAGRLVTTMVVVMIICFLVCWTPYAVMALIVTFGADHLVTPTASVIPSLVAKSSTAYNPIIYVLMNNQ 0 0 FREFLLARLQRVCCRQQAVPRVTPMDDNVHVRLGGEGPSQSQQFLPAGENVENVDMLEYVQENCKPKADSLSTISE* 0 >ENCEPH4_braBel Branchiostoma belcheri (amphioxus) Gt 0...2...0.0 indel x x x x 401 aa 000 nm no_ref genome encephalopsin Amphiop4 introns from braFlo 0 MPLYNTSSGPTQGLPWDTPYSQDPIWNDSSPSNSSEDAVVDQGRGELQDFSDAGYTAIATGLALI 1 2 GLVGSMNNFVVILLIGCHRQLRTPFNLLLLNVSVADLLVSVCGNTLSFASAVQHRWLWGRPGCVWYGFANSLF 1 2 GIVSLVTLSALAFERYCVVVRSSEMLTYKSSLGMIAFIWMYSLLWTSLPLLGWSSYQFEGHS 0 0 VGCSVNWVKHNVNNVSYIITLMVTCFFVPMVVVCWSYACIWRTVRM 0 0 SAEMKSEFGNPQNTGRLVTTMVVVMIVCFLVCWTPYTVMALIVTFGADHLVTPTASVIPSLVAKSSTAYNPIIYVLMNNQ 0 0 FREFLLARLRTFCCRQPRMLRVTPMDDNAHARLVGEGPSHAQQVIPSEENGENVEMRKVQGNQLKADSLSTISE* 0 >ENCEPH5_braFlo Branchiostoma floridae (amphioxus) Gt 0...2...0.0 indel -ZFYVE1 +RTF1 +ATP6V0E1 -Etf1 409 aa 000 nm no_ref genome encephalopsin extra 0 intron 0 MLGMHNVMNATDYDNNNATFAAWNFQRNGTTEEEVEFSGFDTVAVVIAAIGIAGFLSNGAVVLLFLKFRQLRTPFNMLLLNMSVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANHLF 1 2 GLVSLISLAVISYERYRMVVKPKGPGSSYLTYNKVGLAIIFIYLYCLLWTTLPIVGWSSYQLE 0 0 GPKISCSVAWEEHSLSNTSYIVAIFIMCLLLPLLIIIYSYCRLWYKVKK 0 0 GSQNLPPAIRKSSQKEQKIARMVVVMITCFLVCWLPYGAMALVVSFGGESLISPTAAVVPSLLAKSSTCYNPLVYFAMNNQ 0 0 FRRYFQDLLCCGRRLFDASASVNTCNTSAMPRHSPVFQKPDSDQYNGIQKSREPQMRTTGQNAPYRQWIEMQTIAVVVKADEVNNKFGEVKT* 0 >ENCEPH5_braBel Branchiostoma belcheri (amphioxus) Gt 0...2...0.0 indel x x x x 421 aa 000 nm 12435605 AB050609 encephalopsin Amphiop5 extra Nfrag in mrna 0 MLGIYNVVNATEYGNNTTFAAWDFKRNGTGGEEEVEFFGYDAVAGVIAIIGVVGFVSNGAVVVLFLKFPQLRTPFNLLLLNMAVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANHLF 1 2 GLVSLISLAVISFLRYRMVVKPKGPGSSYLTYTKVGLAILFIYLYCLLWTTLPIAGWSSYQLE 0 0 GPKIGCSVAWEEHSWSNTSYIVVLFITCLFAPLLIIVYSYYRLWHKVKQ 0 0 GSRNLPAAMRKSSQKEQKIAMMVIVMITCFMVCWLPYGAMALVVTFGGERLISHTAAVVPSLLAKSSTCYNPVVYFAMNSQ 0 0 FRRYFQDLLCCGRRLFDVSQSVVTGNTAMPRNNSQGFRKDDSDQKQDNGLPKQSEGPMCDHSSNESQMEGSRHNTAASQQWIEMQTIAVVVKAVEVDTSAANEP* 0
>RGR_homSap Homo sapiens (human) ?? 0.2.1.2.1.0.0 indel +PCDH21 -LRIT1 -GRID1 -WAPAL 296 aa 000 nm 17679941 NM_001012720 RGR retinal epithelium Mueller exon-skipping splice isoform 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSQLAWNSAVSLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKREKDRTK* 0 >RGR_galGal Gallus gallus (chicken) ?? 0.2.1.2.1.0.0 indel +PCDH21 -LRIT1 +CHAT -PARG 296 aa 000 nm 14985289 NM_001031216 retinal ganglia RGR 0 MVTSHPLPEGFTEIEVFAIGTALLVE 1 2 ALLGFCLNGLTIISFRKIKELRTPSNLLVLSIALADCGICINAFIAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFLTALASISSSAAVAWDRYHHYCT 1 2 RSKLQWSTAISMMVFAWLFAAFWATMPLLGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYITFLFALSIFNFMIPGFIMMTAYQSIHQKFKKSGHYK 0 0 FNTGLPLKTLVICWGPYCLLSFYAAIENVMFISPKYRM 0 0 IPAIIAKTVPTVDSFVYALGNENYRGGIWQFLTGQKIEKAEVDSKTK* 0 >RGR_xenTro Xenopus tropicalis (frog) ?? 0.2.1.2.1.0.0 indel +PCDH21 -LRIT1 +CHAT -PARG 296 aa 000 nm no_ref BC135113 retinal ganglia RGR 0 MVTSYPLPEGFTETEVFAIGTTLLVE 0 0 ALLGLLLNGLTLLSFYKIRELRTPSNLFIISLAVADTGLCLNAFVAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFVAALSSIGSCAAIAWDRYHQYCT 1 2 RSKLHWSTAVSVVFFIWGFSAFWSAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYISYLFTMAFFEFLVPLFILMTAYQSIYQKMKKSGQIR 0 0 FNTSMPVKSLVFCWGPYCLLCFYAVIQDATILSPKLRM 0 0 IPALLAKTSPAVNAYVYGLGNENYRGGIWQYLTGQKLEKAETDNKTK* 0 >RGR_gasAcu Gasterosteus aculeatus (stickleback) ?? 0.2.1.2.1.0.0 indel +PCDH21 -LRIT1 +CHAT -PARG 296 aa 000 nm no_ref genome retinal ganglia RGR 0 MVSSYPLPDGFTDFDVFSLGSCLLVE 0 0 GLLGILLNAVTIAAFLKVRELRTPSNFLVFSLAVADIGISMNATIAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFVTALASIHFIAAIAWDRYHQYCT 1 2 RTKLQWSSAITLAVFVWLFTAFWSAMPLIGWGEYDYEPLRTCCTLDYTKGDR 2 1 NYVSYLIPMAIFNMAIQVFVVMSSYQSIAQKFKKTGNPR 0 0 FNPNTPLKAMLFCWGPYGILAFYAAVENATLVSTKLRM 0 0 MAPILAKTSPTFNVFLYALGNENYRGGIWQLLTGEKIDVPQIENKSK* 0 >RGR_calMil Callorhinchus milii (elephantfish) ?? 0.2.1.2.1.0.0 indel x x x x 227 aa 000 nm no_ref genome fragment + frag petMar 0 EGFTDFEVFGLGTALLVE 0 0 GLVGLLLNGLTLLAFYKIKELRTPSNLLITSLALSDFGISMNAFIAAFSSFLR 2 1 YWPYGSEGCQTHGFHGFLMALASINACAAIAWDRYHQNCS 1 2 SRLQWSSAITVTVFIWGIAAFWSAMPLLGWGVYDYEPLRTCCTLDYSKGDR 2 1 EFIFPIFIMLSSYQSCKSKFKKTGQVK 0 0 FNTGLPVKTLIFCWGPYSLLCFYATIENITILSPKLRM 0 >PER_homSap Homo sapiens (human) ?? 0.2.0.2.1.0.1 indel -CFI +NOLA1 +EGF -ELOVL6 338 aa 000 nm 17167409 NM_006583 peropsin RRH RRH retinal photoisomerase Retinal epithelium 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDV 1 2 GRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0 >PER_monDom Monodelphis domestica (opossum) ?? 0.2.0.2.1.0.1 indel -CFI +NOLA1 +EGF -ELOVL6 326 aa 000 nm no_ref genome peropsin RRH 0 MFKNNSVKTLAPEKEGPSVFSPIEHKIVAAYLITA 1 2 GVISIVSNVIVLGIFVKYKALRTATNTIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYDGCQ 0 0 IYAGLNIFFGMASIGLLTAVAIDRYLTICQPDL 1 2 GRMTSYNYTLMILTAWVNGFFWALMPIVGWAGYAPDPTGATCTINWRKNDV 2 1 SFVSYTMTVITINFAMPLGVMFYCYYNVSQKMKQYSPSNCPDHINRDWSNQVAVTK 0 0 MSVVMILMFLLAWSPYSIVCLWASFGDPKEIPPAMAIVAPLFAKSSTFYNPCIYVAANKK 2 1 FRRAISAMIRCQTHQSMPISNALPMN* 0 >PER_galGal Gallus gallus (chicken) ?? 0.2.0.2.1.0.1 indel -CFI +NOLA1 +EGF -ELOVL6 335 aa 000 nm 14985289 NM_001079759 peropsin RRH 0 MHWNDSANSSESDAEAHSVFTQTEHNIVAAYLITA 1 2 GVISIFSNIVVLGIFVKYKEFRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYTGCQ 0 0 IYAALNIFFGMASIGLLTVVAVDRYLTICRPDI 1 2 GRRMTTRNYAALILAAWINAVFWASMPTVGWAGYASDPTGATCTANWRKNDV 2 1 PFVSYTMSVIAVNFVVPLTVMFYCYYNVSRTMKQYTSSNCLESINMDWSDQVDVTK 0 0 MSVVMIVMFLVAWSPYSIVCLWSSFGDPKKISPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAILAMVRCQTRQEITISNALPMTVSLSALTS* 0 >PER_xenTro Xenopus tropicalis (frog) ?? 0.2.0.2.1.0.1 indel -CFI +NOLA1 +EGF -ELOVL6 347 aa 000 nm no_ref genome peropsin RRH 0 METLAEVSTLLPAGTGTVNISDASSEVHSVFSQSEHNIVAAYLITA 1 2 GVISILSNIIVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYVGCQ 0 0 IYAGLNIFFGMASIGLLTVVAIDRYLTICRPDIG 1 2 GRRISGRHYTAMILAAWINAVFWSVMPVVGWSSYAPDPTGATCTINWRKNDV 2 1 SFVSYTMSVVAVNFVVPLMVMFYCYYNVSRTMKGYGSRSSLGGINADWSDQTDVTK 0 0 MSMVMIVMFLVAWSPYSIVCLWSSFGDPRKIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAILSMVQCKSRQEVTLDNHFPMNVSQSTLTT* 0 >PER_gasAcu Gasterosteus aculeatus (stickleback) ?? 0.2.0.2.1.0.1 indel +GPR68 -GNPDA1 -ENPEP -C14orf100 338 aa 000 nm no_ref genome peropsin RRH 0 MGIDPEVNVTDDVTLYGGKSAFTQLEHNIVAGYLITA 1 2 GVISLFSNIVVLLMFWKFKELRTATNFIIINLAFTDIGVAGIGYPMSAASDIHGSWKFGYAGCQ 0 0 IYAALNIFFGMASIGLLTVVAIDRYLTICRPDIG 1 2 GQKMTMQSYNLLILAAWLNAVFWSSMPVVGWASYAPDPTGATCTINWRQNDV 2 1 SFISYTMAVIAVNFVLPLSAMFYCYYNVSATVKRYKASNCLDSANIDWSDQMDVTK 0 0 MSIVMIIMFLVAWSPYSIVCLWASFGDPKTIPAPMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAIIGMVRCQTRQRITINSQVPMTTSQQPLTQ* 0 >PER_calMil Callorhinchus milii (elephantfish) ?? 0.2.0.2.1.0.1 indel x x x x 151 aa 000 nm no_ref genome fragment 1 LFVSYTMTVIAVNFVVPLSVMFFCYYNVSKTMSRFISSPSPENINLDWSDQLDVTK 0 0 MSVVMIVMFLLAWSPYSIVCLWASFGNPKLIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRKAIMAMICCQNRQEITINHTLPMTISRVPLTE* 0 >PERa_braFlo Branchiostoma floridae (amphioxus) ?? 0.2.0.2.2.0.0.0 indel x x x x 365 aa 000 nm 12435605 AB050610 peropsin Amphiop3 frag 0 MDIPTETPYGAGDDPAGTGWRWAETDQNGFHKYDHLIVGLYLFVI 1 2 GIIGTVENGITLATFTKFRSLRSPTTMLLVHLAIADLGICIFGYPFSGASSLR 0 0 SHWLFGGVGCQWYGFNGMFFGMANIGLLTCVAVDRYLVICRQDL 1 2 VDKVNYNTYGVMAALGWLFAAFWAALPLVGWAEYSLEPS 1 2 GTACTINWQKNDSLYISYVTSCFILGFALPLAVMMFCYWQ 0 0 ASCFVNKVLKGDISGDLTFPVAVNVDWEYQNHFSK 0 0 MCLAMVAAFVVAWTPYSVLFLFAAFGNPADIPAWITLLPPLIAKSSALYNPIIYIIANRRFRSAIFSMVKGQNPDVE 0 0 TLFARDFRISPIEDTGKEMSSMGNANA* 0 >PERa_braBel Branchiostoma belcheri (amphioxus) ?? 0.2.0.2.2.0.0.0 indel x x x x 365 aa 000 nm 12435605 AB050610 peropsin Amphiop3 0 MDIPTETPYGAEEDIGESAGWRWTETDKNGFHKYDHLIVGLYLFVI 1 2 GIIGTIENGITLATFSKFRSLRSPTTMLLVHLAIADLGICIFGYPFSGASSLR 0 0 SHWLFGGVGCQWYGFNGMFFGMANIGLLTCVAVDRYLVICRHDL 1 2 VDKVNYNTYGVMAALGWLFAAFWAALPLVGWAEYALEPS 1 2 GTACTINFQKNDSLYISYVTSCFVLGFVVPLAVMAFCYWQ 0 ASCFVSKVLKGDIAGDLTFPVAANVDWEYQNHFSK 0 0 MCLAMVAAFVVAWTPYSVLFLFAAFWNPADIPAWLTLLPPLIAKSSALYNPIIYIIANRRFRNAICSMMKGQDPDVE 0 0 DDEHADEHRVRSIEDNDKEIISMVNLNMTV* 0 >PERb_braFlo Branchiostoma floridae (amphioxus) ?? 0.2.0.2.2.1.0.0.0 indel x x x x 522 aa 000 nm 12435605 AB050607 peropsin Amphiop2 PER/NEUR frag 0 MIPTNNNTENNDLEWGLEKEHGVSATIMGVYLTIV 1 2 GLVSTVGNATVVLMFMLKWRQLCRKAPNLLIINLAAVDLCISVFGYPFSASSGFANQWLFSDAICT 0 0 LYGFSCFLLSMVSMHTLCLISAHRYITICRPEH 1 2 ASKLTMTRTILAVVGAWVYGISVAVPPLFGIA 1 2 GYTYESFGLSCTIDFHGTTVADMVYLSILIILCYVINVAVMGTCYFKIIRK 2 1 FSKHRFREVRDVRTSHQHSFERGVTL 0 0 RCILMTLFYLISWTPYTAVAVWTMVGPPPPVQLGMVAALTAKTHCAFNPILYMLMSE 0 0 VYRKLVLRTMCPCCFNKISNKLVRLPADDSKHSGNLDIFTVGYNTRDQAVQINKNAARRFCFVMET 0 >PERb_braBel Branchiostoma belcheri (amphioxus) ?? 0.2.0.2.2.1.0.0.0 indel x x x x 522 aa 000 nm 12435605 AB050607 peropsin Amphiop2 RRH 0 MIPTNNNTENNDLEWGLEKEHGVSATIMGVYLTIV 1 2 GLVATVGNATVVLMFIMKWRQLCRKAPNLLVINLAAANLCITIFGYPFSASSGYAHQWLFPDAICT 0 0 LYGFSCFLLSMVSMHTLCLISAHRYITICRPEH 1 2 ASKLTMNRTVLAVIGTWLYAIAVAVPPLFNIA 1 2 RYTYEPSGLSCTIDFRVTTVADLVYLGSLIVLCYVIHVAVMATCYFKIIRK 2 1 FSRHRFRQVRDIRTSHQRSFEMGVTM 0 0 RCILMTLFYLLSWTPYTAVCIWTMVGPPPPVVVSMAAALIAKTHCAFNPILYAFMSE 0 0 VYRKLVFRTMCPCCFNRISCKFVGTPTGGSKVSANPDIFTVDYNSRDQAVQINKAPSRRFCFVMET 0 0 SEDLGSDDTGLTGHSGLWRSGAEVEGLGGLQVTQSPSVSGSELSLSLLDFLPPKPSGRAVSAKLPSPPALNSERATCPESSQQPSDRPATGLRQYQKGDTTRSSVGDLILTEDD VTNLPPASETWGRKKSENPLSYRQTTRRTFGRSRKHSYIVD* 0 >PERc_braFlo Branchiostoma floridae (amphioxus) Go 0.2.2.2.2.0.0 indel x x x x 391 aa 000 nm 12435605 AB050606 peropsin Amphiop1 RRH no petMar frag 0 MNASPSSWLPSGELFTDSPENSSEWPWTDGPTDTAWHHHQTVDPVTYGGYLASAVYLTIT 1 2 GLIAFVGNIFAIIVFLTEKEFRKKEHNSFALNLAIADLSVCVFAYPSSTIS 1 2 GYAGEWMLGDVGCTIYGFLCFTFSLTSMVTLCAISVYRYIVICKPQY 1 2 AHLLTHRRTNYVILGIWLYALVFSVPPLFGVNRYTYEPI 1 2 ITCSLDWNVQHVGETIYTAAVIIIVYVLNVSIMCFCYFNIIFKSANLKFAALASEKTRTAAKKDIWKTSM 0 0 MCLAMVVSFLIAWTPYAVSSTWDILTEEDLPIIATILPTMFAKSSCMMNPIIYSCCNGKFRQAALKTFSK 0 >PERc_braBel Branchiostoma belcheri (amphioxus) Go 0.2.2.2.2.0.0 indel x x x x 391 aa 000 nm 12435605 AB050606 peropsin Amphiop1 RRH no petMar 0 MNASPSSWLSSGEFFTDSPENSSEWPWTDGPTDTTWRHHQSVDSVSYEGYLASAIYIT1 2 LTGLIAFFGNVITITVFLTEKEFRKKQQNGFVLNLAIADLSVCVFAYPSSAI 1 2 AGYAGRWVLGDVGCTIYGFLCFTFALVSMVTLCVISIYRYILICKPQY 1 2 AHLLTHRRTVYVIIGTWLYALVFTVPPLVGVKRYTYEPM 1 2 QITCSLDWNVQHPGEKAYIAAVLVIVYVLQVLIMCFCYFNIIFKSANLKFAALASEKTKMAAKKDTWKTSV 0 0 MCLTMVVSFLIAWTPYAVSSTWDILSAEDLPIIATILPSLFAKSSCMMNPIIYACCNTKFRQAAVKSFRK 0 0 LCGMCKQKVPLSTPQVVLAMQRNTEFTSTVEPTGQAFPMRVLPSISATHTAL* 0 >PER_sacKol Saccoglossus kowalevskii best hit: PERa_braFlo e = -49 Identities = 97/246 (39%) IIYYFFLLSTGLTIFGMSLSCVSSF GRWLFGKFGCYFHGFAGMLFGLGSIGNLTVISIDRYIITCKRSL 1 2 WSYRHYYALLAVAWSNALFWSMMPLFGWSSYALEPEGTSCTIDWMNNDNQYISYVSCVTVTCFILPCAVMTYDYLAAYMKMVKAGYTLSEETEKPNND 0 0 MCIALVAAFLLSWFPSATVFLWAAFGNPGNIPLSFTGVADAFTKIPAVFNPVIYVALNPEFRKYFGKTIGCRRKRKKPIAVRLNGSEQNVENTI* 0 >NEUR_homSap Homo sapiens (human) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 -MUT 355 aa 000 nm 15774036 NM_181744 neuropsin OPN5 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFR 2 1 LHTVTTVRKSSAVLEIHEEV* 0 >NEUR_monDom Monodelphis domestica (opossum) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 -MUT 352 aa 000 nm no_ref genome neuropsin OPN5 0 MALNHSVSPQDDYIPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIISCFSHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 GTWLKRHHAYICLVIIWAYATFWATMPLAGLGNYAPEPFGTSCTLDWWLAQASVTGQTFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQSSHVLEMKLTK 0 0 AMLICAGFLIAWIPYAVVSVWSAFGQPDSIPVQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCQSGGQKAAKKESLRTYR 2 1 HTVATIRKSSAVSETHQEV* 0 >NEUR_ornAna Ornithorhynchus anatinus (platypus) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 - 351 aa 000 nm no_ref genome neuropsin OPN5 0 MTNYSAPQLGDYLPHYLREGDPFVSKLSWEADLVAGVYLVII 1 2 GVLSTLGNGYVIYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIVSCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 GTWLKRHHAYICLAIIWAYASFWATMPLVGLGNYAPEPFGTSCTLDWWLAQASVAGQAFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQNSHVLEMKLTK 0 0 AMLICAGFLIAWIPYAVVSVWSAFGQPDSIPIQFSVVPTLLAKSAAMYNPIIYQVIDCRISCCRLGGPKTGKKESLKNSR 2 1 HSMSTIRKPSAVSGPHQEV* 0 >NEUR_galGal Gallus gallus (chicken) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 - 352 aa 000 nm no_ref genome neuropsin OPN5 0 MASDCNSSSQEEYLPHYMQQEDPFASKLSREADIIAGFYLTVI 1 2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCRSGGPKTLQKKSSLKES 2 1 YTISSHRDSAALSGTQLEV* 0 >NEUR_anoCar Anolis carolinensis (lizard) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 +ITSN2 340 aa 000 nm no_ref genome neuropsin OPN5 0 MEQGQNISSQDDNQQEEDPFASKLSVEADIVAGVYLLVI 1 2 GILSTLGNGYVIYMSTQRKKKLKPAEIMTVNLAVCDLGIS 1 2 VGKPFSIIAFFSHRWIFGWSGCRWYGWAGFFFGIGSLITMTAVSLDRYFKICHLSY 1 2 GTWLKRHHVFICLGIIWSYAAFWATIPFAGFGNYAPEPFGTSCTLDWWLAQGSVAGQAFILNILFFCLVLPTAVIMFCYVKIIAKVQSSTKEVAHYDTRIQNQHVLEMKLTK 0 0 VAMLICAGFMFAWIPYAVVSVWSAFGRPDSVPIKVSVIPTLLAKSAAMYNPVIYQVIDCKSACCRPGNLQPLQKKNSRYVF 2 1 MLQWDKGHDEV* 0 >NEUR_xenTro Xenopus tropicalis (frog) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 - 340 aa 000 nm no_ref genome neuropsin OPN5 truncated 0 MAGNSSYREESGYIPHYERDSDPFASKLSREADIFAGVYLMAI 1 2 GILSTLGNGYVIYMACSRKKKLRPAEIMTINLAVCDLGIS 1 2 VTGKPFAIVSCFSHRWVFGWNACRWYGWAGFFFGCGSLITLTVVSLDRYLKICHLRY 1 2 GTWLKRRHAFIALAVIWAYATLWATLPLVGVGNYAPEPFGTTCTLDWWLAQASVKGQIFVLSMLFFCLLFPTMVIVFSYAKIIAKVKSSAKEVAHFDTRNQNNHTLEIKLTK 0 0 AMLICAGFLIAWFPYAVVSVWSAFGQPDSIPIELSVVPTMMAKSASMYNPIIYQVIDCKPACCKKDKSLQNTTSRYVFVVYIPFHHYR 2 >NEUR_gasAcu Gasterosteus aculeatus (stickleback) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 - 331 aa 000 nm no_ref genome neuropsin OPN5 truncated 0 MENETWTHPSYIPHYLLRGDPFASRLSKEADIIAAFYICII 1 2 GIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGIS 1 2 VTGKPFFVVSSFAHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 2 GTWLKRQHAFLCLVFVWMYAAFWATMPLVGWGNYAPEPFGTSCTLDWWLAQASVSGQSFVVAILFFCLVLPAGIIVFSYVMIIFKVKSSAKEISNFDARIKNSHNLEIKLTK 0 truncated 0 AMLICAGFLIAWIPYAVVSVVSAFGEPDSVPISVSVIPTLLAKSSAMYNPIIYQVLDLKNSCMKSSCFKGLKKPRHFRKSR 2 >NEUR_calMil Callorhinchus milii (elephantfish) ?? 0.2.2.2.0.1 indel x x x x 209 aa 000 nm no_ref genome fragment maybe petMar 2 GLLSTLGNGYVIYLSITQKRKLKPPEILITNLAISDFGMS 1 2 VGGQPFLIISCFSHRWIFGWVGCRWHGWAGFFFGCGSLITMTVVSLDRYLKICHLQY 1 2 GSWLQRRHVFMSLAFIWFYAAFWATMPLVGWGNYAPEPFGTSCTLDWWLARVSVSGLIFVLTILFFCLLLPIIIIVFSYIKIIAKVKSSAKEVAHFDSRIQNHHSLEMNLTK 0
>MEL1_homSap Homo sapiens (human) Gq 0.0.1.2.2.1.1.1.0.0 indel -GRID1 -WAPAL +LDB3 +BMPR1A 483 aa 000 nm 16961436 NM_033282 melanopsin OPN4 0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGRLPSISPT 0 0 APGTWAAAWVPLPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 1 SRSLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2 1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2 1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2 1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLTSHTSNLSWISIRRRQESLGSESEV 0 0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGK 0 0 TKGLIPSQDPRM* 0 >MEL1_monDom Monodelphis domestica (opossum) Gq 0.0.1.2.2.1.1.1.0.0 indel -GRID1 -WAPAL +LDB3 +BMPR1A 483 aa 000 nm no_ref genome melanopsin OPN4 0 MNPSPMLRGLSCPAQDTNCTKIMASMSEWNNTEEDAYHLVDLPSIAPT 0 0 AVVLPPSSQNIFPTADVPDHAHYTIGATILAVGFTGVLGNLLVIYTFCR 2 1 LRTPANMFIINLAISDFFMSFTQAPVFFASSMYKRWIFGEK 1 2 ACEFYAFCGALFGITSMITLMAIALDRYFVITRPLASIGVISKKKTGFILLGVWLYSLAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYTTFTPSVRAYTMLLFCFVFFIPLIVIIYCYIFIFRAIQDTNK 2 1 AVHSIGSGESTASPRHCQRMKNEWKMAKIALVVILLYVLSWAPYSTVALVAFAG 2 1 YSHILTPYMNSVPAIIAKASAIHNPIIYAISHPKYR 2 1 MAIAQNFPCLRALLCVRHPRTRSFSSYRFTRRSTMTSQASDISWLPRGRRQLSLGSESEI 0 0 GWNNMEAGTTSLTSRNQQGSCRMDQETMETRELAAIAKAKGRSWETLEK 0 0 TLEEMDDSSLLEVSVDMEQ* 0 >MEL1_galGal Gallus gallus (chicken) Gq 0.0.1.2.2.1.1.1.0.0 indel -GRID1 -WAPAL +LDB3 +BMPR1A 529 aa 000 nm 16856781 AY88294 melanopsin OPN4m 0 MDLPPRAPT 0 0 KMTVKDVRGAFPTVDVPDHAHYTIGTVILIVGITGTLGNFLVIYAFCR 2 1 SRTLQKPANIFIINLAVSDFLMSITQSPVFFTNSLHKRWIFGEK 1 2 GCELYAFCGALFGITSMITLMVIALDRYFVITKPLASVRVMSKKKALIILVGVWLYSLAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFIPLIAIIYSYVFIFEAIKKANK 2 1 SVQTFGCKHGNRELQKQYHRMKNEWKLAKIALIVILLYVISWSPYSVVALVAFAG 2 1 YSHVLTPFMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 TAIATYVPCLGFLLRVSPKESRSFSSYPSSRRTTITSQSSETSGLQKGKRRLSSISDSES 0 0 GCTDTETDITSMISRPASSQVSYEMGEDTTQTSDLGGKPKVKSHDSGIFRK 0 0 TVVDADEIPMVEINDTEHSATSTCKTSEKCNVEEIQ 0 0 RSESLSGIGLREGESRHRTSASQIPSIIITYSNVQGVELHSGYSAGFLHPKNKSHKQNKSSNS* 0 >MEL1_xenTro Xenopus tropicalis (frog) Gq 0.0.1.2.2.1.1.1.0.0 indel -GRID1 -WAPAL +LDB3 +BMPR1A 596 aa 000 nm 16856781 DQ384639 melanopsin OPN4m 0 MNYQSVRKGITCPPQDANCSRILESLNSWNNSEVNSYKLVELPPIVTT 0 0 ETPQYEIHHVYPTVDVPDHVHYVVGAVILAVGITGMLGNFLVIYAFCR 2 1 SRSLRSPANMFIINLAITDFLMSVTQAPVFFATSLHKRWIFGEK 1 2 GCELYAFCGALFGITSMITLMVIAVDRYFVITRPLTSIGVMSKKRAVLILSGVWLYSLAWSLPPFFGW 1 2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFCFVFFIPLFIIIYCYIFIFKAIKNTNR 2 1 AVQKIGTDNNKESHKQYQKMKNEWKMAKIALIVILLYVVSWSPYSTVALLAFAG 2 1 YASILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 MAIAKYIPCLGSLLRVKRRDSRSYSSYPSSRRSTVTSHCSQSSDVGGHPKLKNHLPSVSDSES 0 0 GWTDTEADSSVNSRPASRQVSYEMGKDTTETNDLKSKAKLKSHDSGIFEK 0 0 TSMDADDISLVELGTVDRSSPIM 0 0 ANKHLNGLGQRKGDSFTRRSPSSRIPSIVVTHSNHQGSPAAVRHNSTLPGIKVSNSQDREKELKRQIEKVKQYVPIVTITSDTENSTGGFSNELLPANTS* 0 >MEL1_danRer Danio rerio (zebrafish) Gq 0.0.1.2.2.1.1.1.0.0 indel - +USP54 +LDB3 +BMPR1A 594 aa 000 nm no_ref AY078161 melanopsin OPN4m 0 MMSGAAHSVRKGISCPTQDPNCTRIVESLSAWNDSVMSAYRLVDLPPTTTTTTSVA 0 0 MVEESVYPFPTVDVPDHAHYTIGAVILTVGITGMLGNFLVIYAFSR 2 1 SRTLRTPANLFIINLAITDFLMCATQAPIFFTTSMHKRWIFGEK 1 2 GCELYAFCGALFGICSMITLMVIAVDRYFVITRPLASIGVLSQKRALLILLVAWVYSLGWSLPPFFGW 1 2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFIPLIVIIYCYFFIFRSIRTTNE 2 1 AVGKINGDNKRDSMKRFQRLKNEWKMAKIALIVILMYVISWSPYSTVALTAFAG 2 1 YSDFLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 LAIAKYIPCLRLLLCVPKRDLHSFHSSLMSTRRSTVTSQSSDMSGRFRRTSTGKSRLSSASDSES 0 0 GWTDTEADLSSMSSRPASRQVSCDISKDTAEMPDFKPCNSSSFKSKLKSHDSGIFEK 0 0 SSSDVDDVSVAGIIQPDRTLTN 0 0 AGDITDVPISRGAIGRIPSIVITSESSSLLPSVRPTYRISRSNVSTVGTNPARRDSRGGVQQGAAHLSNAAETPESGHIDNHRPQYL* 0 >MEL1D_danRer Danio rerio (zebrafish) Gq 0.0.1.2.2.1.1.1.0.0 indel - +USP54 +LDB3 +BMPR1A 473 aa 000 nm no_ref genome melanopsin OPN4m 0 QVAMVQDVRHPFPTVDVPDHAHYTIGSVILAVGITGMVGNLLVMYAFCK 2 1 SRSLRTPANMFIINLAVTDFLMCVTQTPIFFTTSLHKRWIFGEK 1 2 GCELYAFCGALFGICSMITLMIIAVDRYFVITRPLASIGVMSRKRALLILSAAWAYSMGWSLPPFFGW 1 2 SGAYVPEGLLTSCSWDYMTFSPSVRAYTMLLFTFVFFIPLFVIIYCYFFIFKAIRETNR 2 1 AVGKINGEGGPRDSIKKIHRMKNEWKMAKIALIVILLYVISWSPYSCVALTAF 2 1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 SAIAKYIPCLGVLLCVPRRDRFSSSSFISTRRSTLTSQSSETSSNLHRAGKARLSSVSDSES 0 0 GWTDTEADLSTASSRPASRQVSSEIRKDLCDIKHSSSLRLKVKSRDSGIFDR 0 0 0 0 QNDVSEKADEKRPLVRIPSIIVTSETCPAVLPAGHSSRLIPGAPAVTDS* 0 >MEL1_takRub Takifugu rubripes (teleost) Gq 0.0.1.2.2.1.1.1.0.0 indel - +USP54 +LDB3 +BMPR1A 555 aa 000 nm no_ref genome melanopsin OPN4m 0 MNFGKSALQPPAQQSVVSCGGGGPEPNCTLRLAVTVMMSVRLAELQLHAST 0 0 LQVAMVRPFPTVDVPDHAHYTIGSVILVIGITGMIGNFLVIYAFCR 2 1 SRSLRTPANMFIINLAVTDLLMCVTQTPIFFTTSMYKRWIFGEK 1 2 GCELYAFCGALFGICSMITLTVIAIDRYFVITRPLTSIGVLSRKRAFVILMTVWIYSLGWSLPPFFGW 1 2 SGAYVPEGLLTSCTWDYMTFSPSVRAYTMLLFIFVFFLPLFIIIYCYFFIFRAIRATNK 2 1 AVGKVNGSVHSHSRRRESVKNFQRLQNEWKMAKIALMVILLYVISWSPYSCVALTAFAG 2 1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 LALAKYIPCLGFLLCISPHELQSTSSSFMSLRRSTVTSQTSDISGQFRPQSKPRRSSASDSES 0 0 CLTDTEADLSSMGSRPASRQVSCDISRDTTELPEYKPASSFNSKVKSPDSGIFEK 0 0 TSFDFDASMAASRERSSIPN 0 0 SGEFPEGHVMRRTLARIPSIIITSESSHFLPNGRKASSTTCIANGSDIKVGPR* 0 >MEL1_gasAcu Gasterosteus aculeatus (stickleback) Gq 0.0.1.2.2.1.1.1.0.0 indel - - +LDB3 +BMPR1A 556 aa 000 nm no_ref genome melanopsin OPN4m 0 MNAGESELLLPTQQSILPCGDHEPNCPVAQAETLALSAASANGSA 0 0 VQVAMVSRAPHPYPTVDVPDHAHYTIGSVILAIGITGIIGNVLVIYAFSK 2 1 SRSLRTPANMFIINLAITDLLMCVTQAPIFFTTSMHKRWIFGEK 1 2 GCELYAFCGALFGICSMITLTVIALDRYFVITRPLTSIGMMSRRRALLILMGAWTYSLGWSLPPFFGW 1 2 SGAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFLPLFIIIYCYFFIFRAIRVTNR 2 1 AVGKMNGSIHSHGSGRDSTKNFHRLQNEWKMAKIALIVILLYVVSWSPYSAVALTAFAG 2 1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 IALAKYIPFLGVLLCVPPRELRSASSSFRSTRRSTVTSQTSDVSSQQRRQGSRNSRLSSASDSES 0 0 CLTDTEADGSSVGSRPASRQVSCDIGRDTAELPEFKPSSSFKSKMKSHDSGIFEK 0 0 SYDTDISMAGVSERGSIPN 0 0 QTDFAEGRDRRSTIGRIPSIVITSETSPFLPTGRNGSCNGRPKTANSSHPGAGSG* 0 >MEL1_oryLat Oryzias latipes (medaka) Gq 0.0.1.2.2.1.1.1.0.0 indel - +USP54 +LDB3 +BMPR1A 504 aa 000 nm no_ref genome melanopsin OPN4m 0 LQVAMVPQTFHPFPTVDVPDHAHYTIGSVILAIGITGIIGNFLVIYAFSR 2 1 SRSLRTPANMFIINLAITDLLMCVTQSPIFFTTSMHKRWIFGEK 1 2 GCELYAFCGALFGICSMITLTVIAIDRYFVITRPLTSIGVLSRKRALLILSAAWAYSLGWSLPPFFGW 1 2 SGAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFLPLFIIIYCYVFIFRAIRSTNR 2 1 AVGKINGNTRDAVKSFNRLQNEWKMAKIALIVILLYVISWSPYSTVALTAFAG 2 1 YADMLTPYMNSIPAVIAKASAIHNPIIYAITHPKYR 2 1 MALAKYIPGLGVLLCIHPKDLRSASSSFVSTRRSTVTSQSSDISSQLRRQSTFKSRLSSLSDSES 0 0 GLTDTEADLSSLSSRPASRQVSCEISRDTAELPDFKHTSSFKAKLKNNDSGIFEK 0 0 TSFDTVSIGGVSEHNSIPS 0 0 NRDFGDGNVTRATIGRIPSIVVTSEMSPFLPVGRNGSRTNRSKMANSSAGAGPV* 0 >MEL1_calMil Callorhinchus milii (elephantfish) Gq 0.0.1.2.2.1.1.1.0.0 indel - - - - 369 aa 000 nm no_ref genome melanopsin OPN4m 0 ASVTDAQHHHMFPTVDVPDHAHYIIGATILAVGVTGMVGNFLVIYAFLR 2 1 SRSLRTPANTFIINLAATDFLMSVTQSPIFFITSIHKRWIFGEK 1 2 GCELYAFCGALFGITSMITLMVIALDRYFVITRPLASIGVLSHRRAGLIILSLWLYSLAWSLPPFFGW 1 2 SGAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFCFVFFIPLGVIIYCYIFIFRAIKSTNK 2 1 KVGGSTNRESQKQHQRMKNEWKMAKIALIVILLFVISWSPYSTVALTAFAG 2 1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 MAIAKYVPLLGLLLRVSRRDSRTSGQYYSTRRSTLTSQTSDLSGYPRGKGRLSSASDSES 0 >MEL1b_calMil Callorhinchus milii (elephantfish) Gq 0.0.1.2.2.1.1.1.0.0 indel x x x x 113 aa 000 nm no_ref EB687868 melanopsin OPN4m 1 SKSLRTPANMFIINLAISDFFMSATQPPVFFVTSLHKRWIFGEK 2 GCKLYAFCGALFGITSMITLMAISIDRYWVITKPLQSISSTTTKKNTLKVIILVWLYSLAWSLPPLLGW 1 >MEL1_petMar Petromyzon marinus (lamprey) Gq 0.0.1.2.2.1.1.1.0.0 indel x x x x 205 aa 000 nm no_ref genome fragment 1 SKSLRSPANIFIINLAFADFFMSITQTPIFFVTSLHKRWIFGEK 1 2 GCELYAFCGALFGIASMVTLMVIATDRYLVLTRPLASIGAMSKRRAMYITAAVWFYSLAWSLPPFFGW 1 2 AYVPEGLMTSCTWDYVTFTPAVRSYTMLLFCFVFFIPLIVIIFCYVRIFAAIKNTNR 2 1 YADMLTPYMNSVPAIIAKASAIHNPIVYAITHPKYR 2 >MEL1a_braFlo Branchiostoma floridae (amphioxus) Gq 0.0.1.2.2.1.1.1.0.0 indel - - - - 709 aa 000 nm no_ref genome melanopsin Amphi-mop 12 exons +tandem dup assembly error 0 MTELPSFQPPTNSTEEENAVFPTALTEWISE 0 0 VGNQVGEAALKLLSGEGDGMEVTPTPGCTGNASVCNGTDSGGGVVWDIPPLAHYIVGTAVFCVGCCGMFGNAVVVYSFIK 2 1 SKGLRTPANFFIINLALSDFLMNLTNMPIFAVNSAFQRWLLSDF 1 2 ACELYGFAGGLFGCLSINTLMAISMDRYLVITKPFLVMRIVTKQR 0 0 VMFAILLLWIWSLVWALPPLFGWSAYVPEGF 1 2 GTSCTFDYMTPKLSYHIFTYIIFFTMYFIPMGVIIYCYYNIFATVKSGDKQFGKAVKEMAHEDVKNK 0 0 AQQERQRKNEIKTAKIAFIVITLFLSAWTPYAVVSALGTLGYQDLVTPYLQSIPAVFAKSSAVYNPI 1 2 VYAITHPKFRAAVKKHIPCLSGCLPADEEETKTKTRGATTTASMSMTQTTAPTV 0 0 HDPQASVHSGSSVSVDDSSGVSRQDTMMVK 0 0 VEVDNRMEKAGGGAADTAPKDGTSVPTVSAQIEVRPSGNVNTKAEVIPSPQSAAVAHGASASPVPK 0 0 VAELSSSVSLESAAIPGKIPTPLPSQPIAAPIERHMAAMADDPPPKPRGVATTVNVRRSESGYERSQDSLRKK 0 0 AVSETRSRSFNSTKDHFASERQTSTTLNQPRDMYSGDMVKKTRQSPEKQEYDNPAFDAGIAEIDTDSENETEGSYDMLSVRFQAMAEEPPVETYRKASDMSINLGKASLMLTEAHDETVL* 0 >MEL1a_braBel Branchiostoma belcheri (amphioxus) Gq 0.0.1.2.2.1.1.1.0.0 indel x x x x 707 aa 000 nm 15936279 AB205400 melanopsin Amphi-mop 0 MTEIPSFQPPINATEVEEENAVFPTALTEWFSE 0 0 VGNQVGEVALKLLSGEGDGMEVTPTPGCTGNGSVCNGTDSGGVVWDIPPLAHYIVGTAVFCIGCCGMFGNAVVVYSFIK 2 1 SKGLRTPANFFIINLALSDFLMNLTNMPIFAVNSAFQRWLLSDF 1 2 ACELYGFAGGLFGCLSINTLMAISMDRYLVITKPFLVMRIVTKQR 0 0 VMFAILLLWIWSLVWALPPLFGWSAYVSEGF 1 2 GTSCTFDYMTPKLSYHIFTYIIFFTMYFIPGGVMIYCYYNIFATVKSGDKQFGKAVKEMAHEDVKNK 0 0 AQQERQRKNEIKTAKIAFIVISLFMSAWTPYAVVSALGTLGYQDLVTPYLQSIPAMFAKSSAVYSPI 1 2 VYAITYPKFREAVKKHIPCLSGCLPASEEETKTKTRGQSSASASMSMTQTTAPV 0 0 HDPQASVDSGSSVSVDDSSGVSRQDTMMVK 0 0 VEVDKRMEKAGGGAADAAPQEGASVSTVSAQIEVRPSGKVTTKADVISTPQTAHGLSASPVPK 0 0 VAELGSSATLESAAIPGKIPTPLPSQPIAAPIERHMAAMADEPPPKPRGVATTVNVRRTESGYDRSQDSQRKK 0 0 VVGDTHRSRSFNTTKDHFASEQPAALIQPKELYSDDTTKKMARQSSEKHEYDNPAFDEGITEVDTDSENETEGSYDMLSVRFQAMAEEPPVETYRKASDLAINLGKASLMLSEAHDETVL* 0 >MEL1b_braFlo Branchiostoma floridae (amphioxus) Gq 0.0.1.2.2.1.1.1.0.0 indel - - - - 402 aa 000 nm no_ref genome melanopsin Amphiop6 0 MSPNLTNTSLLPNRTDRPELSPADVTMQLVFGSMMLVFGLIGVVGNAVALYAFCR 2 1 SRSLRRPKNYLIANLCLTDMVVCLVYSPIIVTRSLSHG 2 1 LPSKESCIVEGFVVGLGSIVSICSLAGIAVERYVTITQPIKSLSILTHRALLGAVSAVWVYAFLLAFPPLVGWGRYVSEESKISCTFDYLSTDDATRAHVIVLVIGAFGLPFS VITYCYVRSFATVRKCTKERKQMSPLAKSDSRSEVKAAVNSFVITTSFCLCWCPYAVVATMGVSGFTVHSHAVFIAALLAKLSVLFNPVAYVLSIP 1 2 NSNVNIESTELTVPYSASRESCLLSRAATERLAGRSPSLTDIVREFGLQQTASHRE >MEL1b_braBel Branchiostoma belcheri (amphioxus) Gq 0.0.1.2.2.1.1.1.0.0 indel x x x x 402 aa 000 nm 12435605 AB050611 melanopsin Amphiop6 0 MSSNLTNVSLVANRTDQTELSPTDVTMQLIFGSMMLVFGLIGVVGNVVALYAFCR 2 1 TRSLRRPKNYVVANLCLTDMFVCLVYCPIVVSRSFSHG 2 1 FPSKESCIVEGFMVGVGSIASICSLAAIAVERYLSVTQPLKSLTILTQRKLLVAVLTVWVYSLLLAFPPLVGWGRYVREETYISCTFDYLSTDDATRAYVITLVMGAFGFPLL TIAYCYIRVFTTARKHAEERKFMSPLKRPESRTEIKTAVTACVITTSFCLCWCPYAVVATLGISGVSVQQQTVFSAALLAKLTVIINPIVYVLSIPNFRKALFAQEREKYASED VVLTSLPGKTRRMKKVERSQSSNSNVVIEVKESSMAYSTSRESCLLSRAATKRLAGKTKSIVDLVDEFGLQETAPHKESLV* >MEL2_galGal Gallus gallus (chicken) Gq 0.0.1.2.2.1.1.1.0.0 indel +GRID2+SMARCAD1 -PGDS -SEC24B +COL25A1 544 aa 000 nm 17977531 AY882944 melanopsin 0 MGTQPHSVTKSEIPDHVLYTVGTCVLVIGSIGIIGNLLVLYAFYS 2 1 NKKLRTPQNFFIMNLAVSDFLMSASQAPICFVNSLHREWILGDI 1 2 GCDLYAFCGALFGITSMMTLLAISVDRYLVITKPLRSIQWTSKKRTIQIIAAVWLYSLGW 1 2 SVAPLLGWSSYVPEGLMISCTWDYVTYSPANRSYTMILCCCVFFIPLIIILHCYLFMFLAIRSTGR 2 1 DVQKLGSCSRKSFLSQSMKNEWKLAKIAFVVIIVYVLSWSPYACVTLIAWAG 2 1 RGNTLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYRK 2 1 TIHNAVPCLRFLIRISKNDLLRGSINESSFRTSLSSHQSLAGRTKNTCVSSVSTGEA 0 0 NWSDVELDTVEPAHEKLQPRRSHSFSSSLRQKRDLLPDSYSCSEETEEK 0 0 VSLSSSYLEKVLGRSAFPSSPVALVTSSLRAASLPVGLNSSSASRGAGSDISQMKTEESHNNGGLDSIVSNTVPQIIIIPTSETNLFQEEPEEEETELFHFHDKKNNLLDLEGLSSSTEFLEAVEKFLS* 0 >MEL2_anoCar Anolis carolinensis (lizard) Gq 0.0.1.2.2.1.1.1.0.0 indel +GRID2+SMARCAD1 -ATOH1 +PDLIM5 +BMPR1B 290 aa 000 nm no_ref genome melanopsin 0 MGPHHRTKVDVPDHVLYTVGSCVLVIGCIGITGNLLVLYAFYS 2 1 NKRLRTPPNYFIMNLAVSDFLMSATQAPICFLNSMHKEWVLGDI 1 2 GCNLYAFCGALFGITSMITLLAISVDRYCVITKPLQSIKRTSKKRTCIIIVFVWLYSLGWSVCPLFGW 1 2 SSYIPEGLMISCTWDYVTYSPANRSYTMMLCCCVFFIPLVIIFHCYIFMFLAIRSTGR 2 1 RKSSISHSIKSEWKLAKIAFVAIVVFVLSWSPYACVTLISWAG 2 1 TLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYRK 2 1 TIRSAVPCLRFLIPISKSDLSTSSMSESSFRASVSSRHSFSYRNKSTYISSISAKET 0 0 TWCDVELDPVESGHKKLQAYRSNSFSAKGVAEEESGLLLRTNNCNVPARKK 0 >MEL2_xenLae Xenopus laevis (frog) Gq 0.0.1.2.2.1.1.1.0.0 indel +SMARCAD1 +PDLIM5 +BMPR1B 535 aa 000 nm no_ref genome melanopsin Xmop 21 0 0 0 MDLGKTVEYGTHRQDAIAQIDVPDQVLYTIGSFILIIGSVGIIGNMLVLYAFYR 2 1 NKKLRTAPNYFIINLAISDFLMSATQAPVCFLSSLHREWILGDI 1 2 GCNVYAFCGALFGITSMMTLLAISINRYIVITKPLQSIQWSSKKRTSQIIVLVWMYSLMWSLAPLLGW 1 2 SSYVPEGLRISCTWDYVTSTMSNRSYTMMLCCCVFFIPLIVISHCYLFMFLAIRSTGR 2 1 NVQKLGSYGRQSFLSQSMKNEWKMAKIAFVIIIVFVLSWSPYACVTLIAWAG 2 1 HGKSLTPYSKTVPAVIAKASAIYNPIIYGIIHPKYRE 2 1 TIHKTVPCLRFLIREPKKDIFESSVRGSIYGRQSASRKKNSFISTVSTAET 0 0 VSSHIWDNTPNGHWDRKSLSQTMSNLCSPLLQDPNSSHTLEQTLTWPDDPSPKEILLPSSLKSVTYPIGLESIVKDEHTNNSCVR NHRVDKSGGLDWIINATLPRIVIIPTSESNISETKEEHDNNSEEKSKRTEEEEDFFNFHVDTSLLNLEGLNSSTDLYEVVERFLS* 0 >MEL2_danRer Danio rerio (zebrafish) Gq 0.0.1.2.2.1.1.1.0.0 indel - +FLJ39155 +PDLIM5 - 346 aa 000 nm no_ref genome melanopsin 0 MEPQRQIYKRLDVPDHVHYIIAFLILIIGTLGVSGNALVMFAFYR 2 1 NKKLRSLPNYFIMNLAVSDFLMAITQSPIFFINCLYKEWMFGEL 1 2 GCKIYAFCGALFGITSMINLLAISIDRYLVITKPLQTIQWNSKRRTGLAILCIWLYSLAWSLAPLIGW 1 2 GSYIPEGLMTSCTWDYVSPSPANKSYTMMLCCFVFFIPLSIILYCYLFMFLSVRQASR 2 1 QKSSFVKQQSMRSEWKLAKIAAVVIVVYVLSWAPYACVTLVAWAG 2 1 LTPYSKTLPAVLAKSSAIYNPFIYAIIHNKYRA 2 1 TLAEKVPGLSCLSRSQKDGLSSSTNSDASAQDSSVSRQSSVSKNRLHSTMVQ* 0 >MEL2_tetNig Tetraodon nigroviridis (pufferfish) Gq 0.0.1.2.2.1.1.1.0.0 indel - - - +BMPR1B 404 aa 000 nm no_ref genome melanopsin 0 MEPKDTHITSSFFSKVDVPDHVHYIIAFFVFVIGILGITGNVLVIFAFYS 2 1 NKKLRSLPNYFIVNLAVSDLLMASTQSPIFFINLYKEWMFGET 1 2 ACKMYAFCGALFGITSMINLLAISVDRYVVITKPLQTIRRSSKRRTALAILMVWLYSLAWSLAPLVGW 1 2 GSYIPEGLMTSCTWDYVTYTLANRSYTMMLCCFVFFIPLAIILCCYLLMFLAIRKTSR 2 1 RKSTLIQQKSIRSEWKLAKIAFVVIVVYVLSWSPYACVTLISWAG 2 1 TLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYRK 2 1 TIRSAVPCLRFLIPISKSDLSTSSMSDSSFRSALSCRHSYRSRSTYISSISAKET 0 0 TWCDVELDPVESGHKKLQAYRSNSFSAKGVAEEESGLLLRTNNCNVPARKK 0 >MEL2_gasAcu Gasterosteus aculeatus (stickleback) Gq 0.0.1.2.2.1.1.1.0.0 indel KNTC2 +FLJ39155 +PDLIM5 +BMPR1B 353 aa 000 nm no_ref genome melanopsin 0 MEPDNAHTQRSFINKVDVPDHAHYIVAVFVVVIGTLGITGNALVMLAVYS 2 1 NKKLRNLPNYFIMNLAVSDFLMAFTQSPIFFINCLYKEWAFGET 1 2 GCKIYAFCGALFGIASMINLLAISIDRYLVITKPLQAIHWGSKRRTTLAILLVWLYSLAWSLAPLVGW 1 2 GSYIPEGLMTSCTWDYVTYTLANRSYTMMLCCFVFFIPLGIILYCYLFMFLAIRKTSR 2 1 RKSTLIKQKSMKSEWKLAKIAFVVIVVYVLSWSPYACVTLISWAG 2 1 ILSPYSKAVPAIIAKASAIYNPFIYAIIHNKYRM 2 1 TLAAKFPCLRFLSPTPRKDTSSSISESSYRDSVISRQSTASRTHFITACPDTVN 0 >PIN_stoPur Stronglyocentrotus purpuratus GLEAN3_05569 0.2.2.0.0 16311335 opsin1 PIN-type introns no cdna no sacKow 0 MSNLMTGLVTNVNALSGIGNETPTTIGLSSLVVPVSRTTYNYLTVYTGFLTIFGILNNGIVMILFARFPSLRHPINSFLFNVSLSDLIISCLASPFTFASNFAGRWLFGDLGCTLYAFLVFVA 1 2 GTEQIVILAALSIQRCMLVVRPFTAQKMTHRWALFFISLTWIYSLIICVPPLFGWNRYTYEGPGT 1 2 ACSVAWNSPSPGDTSYIIFIFVLVLVIPFGIIIFCYGLLVYAVKK 0 0 ISRTQAALSSEAKADRKVSKMIFIMILFFLIAWTPYTGFSLYVTFGKNVVITPLAGTFPPFFAKLCTIHNPIIYFLLNKQ 0 0 FKDALIQLFCCGENPFDRDESEHEGRGGRHRHRTAPSATAHIGGRGRASSLPTATSMLDIPQAASTAASSSGKTQNKESLEKGPSTSETTNKRVFELSSKIQKFEISEKNNTPSSSELPGASSLSGALMPPRRAMKNQVGCLPPVDN* 0 >ENCEPH_strPur Stronglyocentrotus purpuratus GLEAN3_03451 modified terminal exon by extending penultimate to stop codon 0 MSLATKKHFIRNAVEEGGHLLEKWDKGG 2 1 YAFIMTFLGLNSLMSHAVIAVDRYLVITKPHF 1 2 GIVVTYPKAFLMISIPWVFSFAWAVFPLAGWGEFTYEGTGAWCSVRWDSDQPQIMSYVLAMMFLTFISSIVIMMYCYICIFLTTRRMPRWATSNSIKTHERNRRRR 2 1 EQKLLKTLIAIAIAFLVAWSPYAITSMIVVFGGSELLSLTATTLPSLFAKSSVMINPIIYAVTSRVFRKSLKK 0 0 MLTSFFPGCMTYIMTDKSPPSSSRPIQLGLCKYHFLY* 0 >MEL1_strPur Stronglyocentrotus purpuratus GLEAN3_22851 opsin4 no cdna losing introns, expressed in larval postoral arm 0 MNAVTTALPHGLNKPTIEAR 2 1 WTKSLRTPPNMLIVNLAISDFGMVITNFPLMFASTIYNRWLFGDA 1 2 GCQFYAFCGALFGIMSIANMTAIALDR 2 1 YYVICWSLEAVRSVTHRRSMIIIIIVWCYAIFWSIPPFFGVGSYVLEGYGLGCTFDFMTKDLNHYLHV SFLFASSFVVPVTIIIVCFTRIAITVRAHRHELNKMRTKLTEDKDKKHKSSIRRANKAKTEFQIAKVGFQVTIFYVLSWM PYSIVAVIGQYFDSDLLTPLGTVVPVIFAKCSAIWNPIIYCLSHEKFNAALKEKLMGMCGIEIPSKHRSMGSQESSVTGR RGMHRQNSSTLSESSVTSTVDQDAIELKDRKQGPATVKVQQEKVEGGTYRRNPGDVTFSKDAGVEVDEKRRGDQGQRDDR VRPQGEGQMDQWSQPPPAPASASAPTPGVNDKEYLTKM* 0 >MEL2_strPur Stronglyocentrotus purpuratus GLEAN3_06737 391 single exon cdna: S.droebachiensis DQ285097 94.7% unpublished light-sensitive tube feet 0 MPTTLMENSTPGWMADDSQMEETHPAFPLIGGYLLVVVLLGTAGNSLVIYTFLRFKKLHSPINLLIVNLSASDLLVATTG TPLSMVSSFYGRWLFGTNACAFYGFVNYYCGCISLNSLAAISVFRYIIVVRGQAQNNKLSLRSSIYAILVIHLYTLIFST PPLYGWNRFVLAGYHTSCDIDFHTKTPLFVSYICYMFFFLFFLPLGLISWSYFKIYQRVSKHSNSMRTSFTGVTKEINSD EKHAWLEKMKTTQILHKPVTFLRLKSSFEPRFKPRFKRRFNHRRTASTLFVTIVVFLFAWFPYCIVSLWVLIGDANSISK LSTTIPSLFAKSSVIYNPLIYVVLNSKFRKALIQTLSFLKCLSKHELSESS* 0 >PER2_strPur Stronglyocentrotus purpuratus Go GLEAN3_27634 overshoots iMet opsin3.2 XM_778236 spread across tandem inline 0 MAASVTESSATEAISRLEPEYMVPLTRTGYLLTAIYLTIV 1 2 GSIATVGNITVICVLCRYRTFRKRSINLLLINMAASDLGVSVAGYPLTTVSGYWGRWLFGDVGCQFYAFCVYTLSCSTISTHAAIAVYRYIYIVKTDL 1 2 RPKLTANFTSGVIVVIWVYAFFWTVTPFVGWSSYIYEPFGTSCSVNWVGRTISDISYMVACTIGVYLLQIFIMLYCYIRVAKK 2 1 IRGVDPGRTEEKDAGVVVFGRLRKREAKIDTHVTK 0 0 MCFMMMLTFIVVWAPYAVECLRAAHVHRISALSSVLPTMFAKSSCMVNPIIFLTSSSKFRQDLGKLWSRPSSQDSLQLEER NKTQRSLYVRHSELGSAHGNDTASVYYEKERIYIGEMRATSIQKEAELLQRDPELLSIASSTNSDVKFVVRDRPKRYTKR PVKPQGPRGPEMFTASGVTNKGSSTSDSGGQSTSSGTTGSKPKRSGRKASRQYSMKSQSEDTGEIFTLDGSALEMMSLRKL* 0 >PER1_strPur Stronglyocentrotus purpuratus Go 17067569 GLEAN3_27633 opsin3.1 RRH no cdna inline tandem partner of PER2_strPur 0 MNSFSEESYVTDPTTTQPTLFLTPLSQTGYLLTALYLTLV 1 2 GIVSTIGNITVLCVLCRYGTFRKRSVNILLMNMAVSDLGVSVAGYPLTAISGYRGRWVFADIGCQFSGFCVYALSCSTISTHAVVAIYRYIYIVKPYH 1 2 RPRLSSSTSCLAILCIWTFTLFWTITPFFGWSSYTYEPFGTSCSINWYGKSLGDLTYIICCVVFVFILPIIIMLYCYIGVAKK 2 1 IKGIDPLRTEERDIAVVFGRLRKHETKIDTRVTK 0 0 ICFMMMASFIVVWTPYAVGSIWASKIGKISASASVLPTMFAKSSCMINPIIFLTSSSKFRADLGKLWNRPSSLEHTIRVEERSREQRSFF VRQSALPDAMVSRSASVYYDKERIYIGEMRAASIQKEADLLHRDPEAISIASSTSSSLQFVLKDRQNRYKKKAGEASKKGSNILHFPYDDTE GSMINNLMRPRSHSVTSDNISRVFAPSLKRPTKKRSMSHPDIPSTSADIFTVSPTTIKNLQKQ* 0
>CILL2_plaDum Platynereis dumerilii (ragworm) Gt 0...2...0.0 indel x x x x 310 aa 000 nm 16311335 CT030681 proto cilliary htgs new 5 exons 1 missing 0 MDDLGFLGNSSVNYTVPLLQEDPLLLRILYFGPTSYVITAIYLCIVGVIGTLSNGVIMYLYFKDKSLRSPMNLLFVNLAMSDFTVAFFGAMFQFGLTCTRKYMSPGMALCDFYGFITFLG 1 2 GLASEMNLFIISVERYLAVVRPFDVGNLTNRRVIAGG 1 2 VFVWLYSLVFAGGPLVGWSSYRPEGLGTWCSISWQDRSMNTMSYVTAVFLGCYFFPVSIIIFCYFNVWRKVKE 0 0 AADAQGAGTAGKAEKSIFRMSVIMVTCYLTAWTPYAIVCLIASYGPPNGLPIYAEVLPSLFAKSSQVYNPIIYVLMNKP 0 0 0* 0 >CILL1_plaDum Platynereis dumerilii (ragworm) Gt 0...2...0.0 indel x x x x 355 aa 000 nm 15514158 AY692353 lophotrochozoa ciliary polychaeta new genomic 0 MDGENLTIPNPVTELMDTPINSTYFQNLNAETDGGNHYIYNAFTATDYNICAAYLFFIACLGVSLNVLVLVLFIKDRKLRSPNNFLYVSLALGDLLVAVFGTAFKFIITARKTLLREEDGFCKWYGFITYLG 1 2 GLAALMTLSVIAFVRCLAVLRLGSFTGLTTRMGVAAMA 1 2 FIWIYSLAFTLAPLLGWNHYIPEGLATWCSIDWLSDETSDKSYVFAIFIFCFLVPVLIIVVSYGLIYDKVRK 0 0 VAKTGGSVAKAEREVLRMTLLMVSLFMLAWSPYAVICMLASFGPKDLLHPVATVIPAMFAKSSTMYNPLIYVFMNKQ 0 0 FRRSLKVLLGMGVEDLNSESERATGGTATNQVAAT*
>LOPH_RHO_plaDum Platynereis dumerilii (polychaete) Gq 0.0.1.2.2.1.1.1.0.0 indel x x x x 383 aa 000 nm 11874910 AJ316544 rhabdomeric melanopsin unavailable genomically 0 MSRSEVLVPGSMSLDGLLTTAHPIGNDSI 0 0 ETILHPYWQQFDIENTIPDSWHYAVAAWMTFFGILGVSGNLLVVWTFLK 2 1 TKSLRTAPNMLLVNLAIGDMAFSAINGFPLLTISSINKRWVWGKL 1 2 WRELYAFVGGIFGLMSINTLAWIAIDRFYVITNPLGAAQTMTKKRAFIILTIIWANASLWALAPFFGW 1 2 GAYIPEGFQTSCTYDYLTQDMNNYTYVLGMYLFGFIFPVAIIFFCYLGIVRAIFAHHA 2 1 EMMATAKRMGANTGKADADKKSEIQIAKVAAMTIGTFMLSWTPYAVVGVFGMIK 2 1 PHSEMFIHPLLAEIPVMMAKASARYNPIIYALSHPKFR 2 1 AEIDKHFPWLLCCCKPKPKAQLPSSTTKGSIASKTEADTSV* 0 >MEL_helRob Helobdella robusta (leech) fragmentary model from scaffold_39 1 TPILRTHANVLIINLALCDLIFSSLIGFPMTALSCFKRHWIWGDL 1 2 GCDFYGFVAGWTGLGSITCLAFISIDRYMAIVHPFYMLNKKSSSVLTLLGCDFYGFVAGWTGLGSITCLAFISIDRYMAIVHPFYMLNKKSSSVLTLLIIVTSYIGIVIEVTKS 1 1 KELKTAKVLACCFGAFLICWTPYAIVAQLGINGFAHLVTPFTSEVPVLFAKTSSIWNPLIYALSHPRYRRAV 0 >MOLL_RHO_lolSub Loligo subulata Z49108 499 Mollusca Cephalopoda complete NETWWYNPYMDIHSHWKQFDQVPAAVYYSLGIFIAICGIIGCAG NGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPMMTISCFLKHWVFGQAACK VYGLIGGIFGLTSIMTMTMISIDRYNVIRRPMSASKKMSHRKAFIMIVFVWIWSTIWA IGPIFGWGAYQLEGVLCNCSFDYITRDASTRSNIVCMYIFAFMFPIVVIFFCYFNIVM SVSNHEKEMAAMAKRLNAKELRKAQAGASAEMKLAKISIVIVTQSLLSWSPYAIVALL AQFGPIEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAIASNFPWILTCCQYDE KEIEDDKDAEAEIPAAEQSGGESVDAAQMKEMMAMMQKMQAQQQQQPAYPPQGYPPQG YPPPPPQGYPPQGYPPQGYPPQGYPPPPQGPPPQGPPPQAAPPQGVD >MOLL_RHO_sepOff Sepia officinalis Go? AF000947 492 Mollusca Cephalopoda complete MGRDIPDNETWWYNPTMEVHPHWKQFNQVPDAVYYSLGIFIGIC GIIGCTGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFIKKWV FGMAACKVYGFIGGIFGLMSIMTMSMISIDRYNVIGRPMAASKKMSHRRAFLMIIFVW MWSTLWSIGPIFGWGAYVLEGVLCNCSFDYITRDSATRSNIVCMYIFAFCFPILIIFF CYFNIVMAVSNHEKEMAAMAKRLNAKELRKAQAGASAEMKLAKISIVIVTQFLLSWSP YAVVALLAQFGPIEWVTPYAAQLPVMFAKASAIHNPLIYSVSHPKFREAIAENFPWII TCCQFDEKEVEDDKDAETEIPATEQSGGESADAAQMKEMMAMMQKMQQQQAAYPPQGA YPPQGGYPPQGYPPPPAQGGYPPQGYPPPPQGYPPAQGYPPQGYPPPQGAPPQGAPPQ AAPPQGVDNQAYQA >MOLL_RHO_todPac Todarodes pacificus Go? X70498 480 Mollusca Cephalopoda complete MGRDLRDNETWWYNPSIVVHPHWREFDQVPDAVYYSLGIFIGIC GIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWI FGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVW LWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFF CYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGANAEMRLAKISIVIVSQFLLSWSP YAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVL TCCQFDDKETEDDKDAETEIPAGESSDAAPSADAAQMKEMMAMMQKMQQQQAAYPPQG YAPPPQGYPPQGYPPQGYPPQGYPPQGYPPPPQGAPPQGAPPAAPPQGVDNQAYQA >MOLL_RHO_entDof Enteroctopus dofleini Go? X07797 475 Mollusca Cephalopoda complete MVESTTLVNQTWWYNPTVDIHPHWAKFDPIPDAVYYSVGIFIGV VGIIGILGNGVVIYLFSKTKSLQTPANMFIINLAMSDLSFSAINGFPLKTISAFMKKW IFGKVACQLYGLLGGIFGFMSINTMAMISIDRYNVIGRPMAASKKMSHRRAFLMIIFV WMWSIVWSVGPVFNWGAYVPEGILTSCSFDYLSTDPSTRSFILCMYFCGFMLPIIIIA FCYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGASAEMKLAKISMVIITQFMLSWS PYAIIALLAQFGPAEWVTPYAAELPVLFAKASAIHNPIVYSVSHPKFREAIQTTFPWL LTCCQFDEKECEDANDAEEEVVASERGGESRDAAQMKEMMAMMQKMQAQQAAYQPPPP PQGYPPQGYPPQGAYPPPQGYPPQGYPPQGYPPQGYPPQGAPPQVEAPQGAPPQGVDN QAYQA >MOLL_MEL_aplCal Aplysia californica (sea hare) Gq-coupled 4 exons melanopsin AASC01108363 uncertainties 0 MNVSSSLTSQPYHELLHPHWLEHEEAPEGVHLSVGVFITLVGVLAVCGNSLVIITCIR 2 1 FKDLRTRSNILIINLAVGDLLMCLIDFPLLAAASFYGEWPYGRQ 1 2 VCQMYAFLTAIAGLVTINTLAVIAADRYWAVVRRPTPGQKLPKCVTSIAVASVWAYSISWALCPILGWGAYVLDGIRTTCTFDFLTRTWENRSFVIGMMIGNFVLPFALMVFSYFRIWVAVRKVKSG 2 1 NVFCAIRHNYNLALGSTLFVKQHRYRLHCEQKTVKIIMFLLIAFTVSWSPYLAVSIIGLFGDRSQLTYQNTLTASLIAKTSMVFNPILYSISHPKVRKRIANLACCYSVRRHQQQTSRIKTGRRSTSSATPSRS* 0 >MOLL_MEL_lotGig most like MOLL_RHO_entDof e-60 84/222 (37%) 338 aa Gq-coupled 0 MSIASHVWTNSSTNHFNFSVLHQHWQNQTPLSTACQYTIGIFISTVAVIAVIGNSIVIWAHVR 2 1 IKSLSTTSNMLILNLCVGCLIMCIVDFPLYATSSFLQKWIFGHK 1 2 VCEIYATITGTAGLLIMNSYSAIAFDRFITVTRYNNPNYPRSKSATMCISGFVWIYSLSWSMAPVVGWSRYQLDGSGTT CTFDYLSTTWTNRSFILSIAFFNFVLPLCFILFAYSRILHLISSHSREMKSYRSAVIISKGKASIPKRFRSERKTAITLLI TVVVFCLSWVPYVIIALIGQFGNQSFITPQISVIPQLVAKLSTVTNPILYSLSHPVVRNKLFLRLRHELYRRPSDSVSSSRGIQMKNIEFI* 0 >MOLL_PERc_patYes Patinopecten yessoensis Go 9287291 AB006455 scop2 MPFPLNRTDTALVISPSEFRIIGIFISICCIIGVLGNLLIIIVFAKRRSVRRPINFFVLNLAVSDLIVALLGYPMTAASAFSNRWIFDNIGCKIYAFLCFNS GVISIMTHAALSFCRYIIICQYGYRKKITQTTVLRTLFSIWSFAMFWTLSPLFGWSSYVIEVVPVSCSVNWYGHGLGDVSYTISVIVAVYVFPLSIIVFSYGMILQEKVCKDSRKN GIRAQQRYTPRFIQDIEQRVTFISFLMMAAFMVAWTPYAIMSALAIGSFNVENSFAALPTLFAKASCAYNPFIYAFTNANFRDTVVEIMAPWTTRRVGVSTLPWPQVTYYPRRRTS AVNTTDIEFPDDNIFIVNSSVNGPTVKREKIVQRNPINVRLGIKIEPRDSRAATENTFTADFSVI* >MOLL_MEL_patYes Patinopecten yessoensis Gq 9287291 AB006454 scop1 49% MOLL_RHO_entDof then MEL scallop retina MADNKSTLPGLPDINGTLNRSMTPNTGWEGPYDMSVHLHWTQFP PVTEEWHYIIGVYITIVGLLGIMGNTTVVYIFSNTKSLRSPSNLFVVNLAVSDLIFSAVNGFPLLTVSSFHQKWIFGSLFCQLYGFVGGVFGLMSINTLTAISIDRYVVITKPLQA SQTMTRRKVHLMIVIVWVLSILLSIPPFFGWGAYIPEGFQTSCTFDYLTKTARTRTYIVVLYLFGFLIPLIIIGVCYVLIIRGVRRHDQKMLTITRSMKTEDARANNKRARSELRI SKIAMTVTCLFIISWSPYAIIALIAQFGPAHWITPLVSELPMMLAKSSSMHNPVVYALSHPKFRKALYQRVPWLFCCCKPKEKADFRTSVCSKRSVTRTESVNSDVSSVISNLSDS TTTLGLTSEGATRANRETSFRRSVSIIKGDEDPCTHPDTFLLAYKEVEVGNLFDMTDDQNRRDSNLHSLYIPTRVQHRPTTQSLGTTPGGVYIVDNGQRVNGLTFNS*
>ENCEPH_apiMel Apis mellifera (bee) Gt 0...2...0.0 indel x x x x 329 aa 000 nm 16291092 NM_001039968 encephalopsin ciliary Gt pteropsin clock 0 MSLNRSTMEHVIYEDQVSPVMYIGAAIALGFIGFFGFTANLLVAIVIVKDAQILWTPVNVILFNLV 0 0 FGDFLVSIFGNPVAMVSAATGGWYWGYKMCLW 2 1 YAWFMSTLGFASIGNLTVMAVERWLLVARPMQALSIR 2 1 HAVILASFVWIYALSLSLPPLFGWGSYGPEAGNVSCSVSWEVHDPVTNSDTYIGFLFVLGLIVPVFTIVSSYAAIVLTLKKVRKRA 1 2 GASGRREAKITKMVALMITAFLLAWSPYAALAIAAQYFN 0 0 AKPSATVAVLPALLAKSSICYNPIIYAGLNNQFSRFLKKIFDARGSRTAVPDSQHTALTALNRQEQRK* 0 >ENCEPH1_anoGam Anopheles gambiae (mosquito) Gt 0...2...0.0 indel x x x x 461 aa 000 nm no_ref XM_312503 encephalopsin GPROP11 adjacent head-to-head tandem GPROP12 0 MYDVTDAAAINSDHQELMAPWAYNGAAVTLFFIGFFGFFLNIFVIALMYKDVQ 0 0 LWTPMNIILFNLVCSDFSVSIIGNPLTLTSAISHRWLYGKSICVAYGFFMSLL 1 2 GIASITTLTVLSYERFCLISRPFAAQNRSKQGACLAVLFIWSYSFALTSPPLFGWGAYVNEAANI 1 2 SCSVNWESQTANATSYIIFLFIFGLILPLAVIIYSYINIVLEMRK 0 0 NSARVGRVNRAERRVTSMVAVMIVAFMVAWTPYAIFALIEQFGPPELIGPGLAVLPALVAKSSICYNPIIYVGMNTQ FRAAFWRIRRSNGVAGQPDSNNTNNSNRDKESARHTAKEGL ECSLDFCHWTVRGTRVSISSAERNVPAPAARERSGGHSVTGSREESRDRHVTLKTMLSVGPRSPSSVAPVAADCSTTDVPTSGDGSVRIVRQDSELSVIHDGGGGGGGSSSRVLVIKSQKPRSNML* 0 >ENCEPH2_anoGam Anopheles gambiae (mosquito) Gt 0...2...0.0 indel x x x x 434 aa 000 nm no_ref XM_312502 encephalopsin GPROP12 0 MNDAPNDVAASAVDYEDLMAPWAYNASAVTLFFIGFFGFFLNLFVIALMCKDMQ 0 0 LWTPMNIILFNLVCSDFSVSIIGNPLTLTSAISHRWIFGRTLCVAYGFFMSLL 1 2 GITSITTLTVLSYERYCLISRPFSSRNLTRRGAFLAIFFIWGYSFALTSPPLFGWGAYVQEAANI 1 2 SCSVNWESQTKNATTYIIFLFVFGLVVPLIVIVYSYTNIIVNMRE 0 0 NSARVGRINRAEQRVTSMVAVMIVAFMVAWTPYAIFALIEQFGPPELIGPGLAVLPALVAKSSICYNPIIYVGMNTQ FRAAFSRVRNKGQQAAADQNTTTMQRELTKSSRDMVECSF DFCRKKSRFKISLVKPTAPLAVVDVSSTSHRDKGTSRSPLDQTVLNETNEDVGRERSGGGGGGGAYAGTRFVRPDFELSVINSGKSILIKSKNFRSNLL* 0
>CHEL_LWS_limPol Limulus polyphemus L03781 520 Arthropoda Chelicerata lateral_eye complete genFut MANQLSYSSLGWPYQPNASVVDTMPKEMLYMIHEHWYAFPPMNP LWYSILGVAMIILGIICVLGNGMVIYLMMTTKSLRTPTNLLVVNLAFSDFCMMAFMMP TMTSNCFAETWILGPFMCEVYGMAGSLFGCASIWSMVMITLDRYNVIVRGMAAAPLTH KKATLLLLFVWIWSGGWTILPFFGWSRYVPEGNLTSCTVDYLTKDWSSASYVVIYGLA VYFLPLITMIYCYFFIVHAVAEHEKQLREQAKKMNVASLRANADQQKQSAECRLAKVA MMTVGLWFMAWTPYLIISWAGVFSSGTRLTPLATIWGSVFAKANSCYNPIVYGISHPR YKAALYQRFPSLACGSGESGSDVKSEASATTTMEEKPKIPEA >CHEL_LWS_ixoSca Ixodes scapularis ocellar TC19272 UP|OPSO_LIMPO (P35361) (57%) 0 MGSEGQRTNMSLLDELASPYMKNGTLVESVPDEMLYMVHPHWYNFKPMNPLWHSLLGFAMVILGVISVVGNSMVIYIMTTSKSLRSPTNMLVVNLAFSDW 2 1 CMMAFMMPTMAANCFAETWILGPFMCEVYGMVGSLFGCGSIWSMVMITLDRYNVIVRGVAAAPLTHKRAALMIFFVWFWALTWTLLPFFGWSR 2 1 YVPEGNMTSCTIDYLTKALWSASYVVAYAGGVYWTPLFINIYCYSKIVRAVAQHEKQLRLQARKMNVASLRANAEQTKTSAEARLAK 0 0 IALMTVGLWFMAWTPYLTIAWAGIFSDGSKLTPLATIWGSVFAKANACYNPIVYGISHPKYRAALARRFPSLVCMPPGGDQLDTRSEASGITTIEDKVMTTET* 0 >INSE_LWS_apiMel Apis mellifera (bee) Gq 0.1.0.0.1.0.0.1 indel x x x x 386 aa 000 nm 16291092 NM_001077825 rhabdomeric Lop2 long wavelength ocelli 0 MDTLNITTSFFIEVMPSNISTLTTTGPQFARQLMRFNNQTVVSKVPEEMLHLIDLYW 2 1 YQFPPLDPLWHKILGLVMIILGIMGWCGNGVVVYVFIMTPSLRTPSNLLVVNLAFSDFIMMGFMCPPMVICCFYETW 0 0 VLGSLMCDIYAMVGSLCGCASIWTMTAIALDRYNVIVK 0 0 GMSGTPLTIKRAMLQILGIWLFGLIWTILPLVGWNR 2 1 YVPEGNMTACGTDYLSQDWTFKSYILVYSFFVYYTPLFTIIYSYYFIVS 0 0 AVAAHEKAMKEQAKKMNVTSLRSGDNQNTSAEAKLAK 0 0 VALTTISLWFMAWTPYLVINYIGIFNRSLITPLFTIWGSLFAKANAIYNPIVYGIS 2 1 HPKYRAALKEKLPFLVCGSTEDQTAATAGDKASEN* 0 >INSE_LWS_papXut Papilio xuthus AB007424 520 Arthropoda Insecta Rh2 complete MAIANLEPGMGASEAWGGQAAAFGSNQTVVDKVTPDMMHLIDPH WYQFPPMNPMWHGLLGFTIGVLGFISITGNGMVVYIFTSTKSLKTPSNLLVVNLAFSD FLMMLCMAPPMLINCYYETWVFGPLACELYACAGSLFGSISIWTMTMIAFDRYNVIVK GIAAKPMTINGALLRILGIWLFSLAWTIAPMLGWNRYVPEGNMTACGTDYLSKSWLSR SYILVYSIFVYYTPLLLIIYSYFFIVQAVAAHEKAMREQAKKMNVASLRSSEAANTSA ECKLAKVALMTISLWFMAWTPYLVINYTGVFETAPISPLATIWGSVFAKANAVYNPIV YGISHPKYRAALYQKFPSLACQPSAEETGSVASGATTACEEKPSA >INSE_LWS_manSex Manduca sexta L78080 520 Arthropoda Insecta White complete genFut MDPGPGLAALQAWAAKSPAYGAANQTVVDKVPPDMMHMIDPHWY QFPPMNPLWHALLGFTIGVLGFVSISGNGMVIYIFMSTKSLKTPSNLLVVNLAFSDFL MMCAMSPAMVVNCYYETWVWGPFACELYACAGSLFGCASIWTMTMIAFDRYNVIVKGI AAKPMTSNGALLRILGIWVFSLAWTLLPFFGWNRYVPEGNMTACGTDYLSKSWVSRSY ILIYSVFVYFLPLLLIIYSYFFIVQAVAAHEKAMREQAKKMNVASLRSSEAANTSAEC KLAKVALMTISLWFMAWTPYLVINYTGVFESAPISPLATIWGSLFAKANAVYNPIVYG ISHPKYQAALYAKFPSLQCQSAPEDAGSVASGTTAVSEEKPAA >INSE_LWS_bomTer Bombus terrestris AY485301 529 Arthropoda Insectapartial genNow YQFPPLNPMWHGILGFVIGLLGFISVSGNGMVVYIFLSTKSLRT PSNMFVINLAISDFLMMFCMSPPMVINCYYETWVLGPLFCQVYAMLGSLFGCGSIWTM TMIAFDRYNVIVKGLSGKPLTINGALLRILGIWLFSLIWTIAPMFGWNRYVPEGNMTA CGTDYFSKDIVSVSYILLYSIWVYFFPLFLIIWSYWFIXQAVAAHEKNMREQAKKMNV ASLRSSENQNTSAECKLAKVALMTISLWFMAWTPYLVINWSGIFSLVKISPLYTIWGS LFAKANAV >INSE_LWS_apiMel Apis mellifera U26026 529 Arthropoda Insecta 540 complete genNow MIAVSGPSYEAFSYGGQARFNNQTVVDKVPPDMLHLIDANWYQY PPLNPMWHGILGFVIGMLGFVSAMGNGMVVYIFLSTKSLRTPSNLFVINLAISNFLMM FCMSPPMVINCYYETWVLGPLFCQIYAMLGSLFGCGSIWTMTMIAFDRYNVIVKGLSG KPLSINGALIRIIAIWLFSLGWTIAPMFGWNRYVPEGNMTACGTDYFNRGLLSASYLV CYGIWVYFVPLFLIIYSYWFIIQAVAAHEKNMREQAKKMNVASLRSSENQNTSAECKL AKVALMTISLWFMAWTPYLVINFSGIFNLVKISPLFTIWGSLFAKANAVYNPIVYGIS HPKYRAALFAKFPSLACAAEPSSDAVSTTSGTTTVTDNEKSNA >INSE_LWS_catBom Cataglyphis bombycinus U32501 510 Arthropoda Insecta complete MMSIASGPSHAAYTWTAQGGGFGNQTVVDKVPPEMLHLVDAHWY QFPPMNPLWHAILGFVIGILGMISVIGNGMVIYIFTTTKSLRTPSNLLVINLAISDFL MMLSMSPAMVINCYYETWVLGPLVCELYGLTGSLFGCGSIWTMTMIAFDRYNVIVKGL SAKPMTINGALLRILGIWFFSLGWTIAPMFGWNRYVPEGNMTACGTDYLTKDLLSRSY ILVYSFFCYFLPLFLIIYSYFFIIQAVAAHEKNMREQAKKMNVASLRSAENQSTSAEC KLAKVALMTISLWFMAWTPYLVINYAGIFETVKINPLFTIWGSLFAKANAVYNPIVYG ISHPKYRAALFQRFPSLACSSGPAGADTLSTTTTVTEGTEKPAA >INSE_LWS_pieRap Pieris rapae AB177984 540 Arthropoda Insecta complete MAITNLDPAPGVAAMQSFGIHAEAFGSNQTVIDKVLPEMMHLID PHWYQFPPLNPLWHALLGFTISVLAFISITGNGMVVYIFTTTKSLKTPSNLLVVNLAF SDFLMMAMMAPPLVVNSYNETWVFGPTACQFYACFGSLFGCVSIWTMTAIAFDRYNVI VKGIAAKPMTINSALLRILGVWLFSLAWTLAPIFGWSRYVPEGNMTACGTDYLSKDWA SRSYIILYAIACYFLPLFLIVYSYWFIVQAVAAHERAMREQAKKMNVASLRSSEQANT SAECKLAKVALMTISLWFMAWTPYLVINFAGVFETSPISPLSTIWGSVFAKANAVYNP IVYGISHPKYRAALYQRFPALACQPSPAEETGSVASAATACTEEKPSA >INSE_LWS_vanCar Vanessa cardui AF385333 530 Arthropoda Insecta complete MAITSLDPGAAALQAWGGQMAAFGSNETVVDKVLPDMLHLVDPH WYQFPPMNPLWHGLLGFVIGILGFISITGNGMVIYIFTTTKSLKTPSNILVVNLAFSD FLMMCVMSPPMVVNCYTETWVFGPLACQLYACAGSLFGCASIWTMTMIAFDRYNVIVK GIAAKPLTINGAMLRVLGIWVFSLAWTVAPLFGWGRYVPEGNMTACGTDYLDKSWFNR SYILIYSIFCYFSPLFLIIYSYFFIVQAVAAHEKAMREQAKKMNVASLRSSDAANTSA ECKLAKVALMTISLWFMAWTPYLVINYAGIFETATITPLATIWGSVFAKANAVYNPIV YGISHPKYRAALYARFPALACQPSPEDNASVASAATATEEKPSA >INSE_LWS_helSar Heliconius sara AF126753 550 Arthropoda Insectapartial HQFPPMNPLWHGLLGFVIGVLGFISVTGNGMVVYIFTTTKSLKT PSNILVVNLAFSDFLMMFMMAPPMVINCYNETWVFGPLACQLYACAGSLYGCVSIWTM TMIAFDRYNVIVKGIAAKPMTINGALLRVFGIWAFSLAWTIAPLFGWGRYVPEGNMTA CGTDYFDQSFSNRSYILLYSIACYYAPLFLIIYSYFFIVQAVAAHEKAMREQAKKMNV ASLRSSDAANTSAECKLAKVALMTISLWFMAWTPYLVINYAGIFKTMT >INSE_LWS_schGre Schistocerca gregaria X80071 520 Arthropoda Insecta complete MASASLISEPSFSAYWGGSGGFANQTVVDKVPPEMLYLVDPHWY QFPPMNPLWHGLLGFVIGVLGVISVIGNGMVIYIFSTTKSLRTPSNLLVVNLAFSDFL MMFTMSAPMGINCYYETWVLGPFMCELYALFGSLFGCGSIWTMTMIALDRYNVIVKGL SAKPMTNKTAMLRILFIWAFSVAWTIMPLFGWNRYVPEGNMTACGTDYLTKDWVSRSY ILVYSFFVYLLPLGTIIYSYFFILQAVSAHEKQMREQRKKMNVASLRSAEASQTSAEC KLAKVALMTISLWFFGWTPYLIINFTGIFETMKISPLLTIWGSLFAKANAVFNPIVYG ISHPKYRAALEKKFPSLACASSSDDNTSVASGATTVSDEKSEKSASA >INSE_LWS_droMel Drosophila melanogaster Z86118 508 Arthropoda Insecta Rh6 complete genNow MASLHPPSFAYMRDGRNLSLAESVPAEIMHMVDPYWYQWPPLEP MWFGIIGFVIAILGTMSLAGNFIVMYIFTSSKGLRTPSNMFVVNLAFSDFMMMFTMFP PVVLNGFYGTWIMGPFLCELYGMFGSLFGCVSIWSMTLIAYDRYCVIVKGMARKPLTA TAAVLRLMVVWTICGAWALMPLFGWNRYVPEGNMTACGTDYFAKDWWNRSYIIVYSLW VYLTPLLTIIFSYWHIMKAVAAHEKAMREQAKKMNVASLRNSEADKSKAIEIKLAKVA LTTISLWFFAWTPYTIINYAGIFESMHLSPLSTICGSVFAKANAVCNPIVYGLSHPKY KQVLREKMPCLACGKDDLTSDSRTQATAEISESQA >INSE_MWS_droMel Drosophila melanogaster X65877 478 Arthropoda Insecta Rh1 complete genNow MDSFAAVATQLGPQFAAPSNGSVVDKVTPDMAHLISPYWDQFPA MDPIWAKILTAYMIIIGMISWCGNGVVIYIFATTKSLRTPANLLVINLAISDFGIMIT NTPMMGINLYFETWVLGPMMCDIYAGLGSAFGCSSIWSMCMISLDRYQVIVKGMAGRP MTIPLALGKIAYIWFMSTIWCCLAPVFGWSRYVPEGNLTSCGIDYLERDWNPRSYLIF YSIFVYYIPLFLICYSYWFIIAAVSAHEKAMREQAKKMNVKSLRSSEDADKSAEGKLA KVALVTISLWFMAWTPYLVINCMGLFKFEGLTPLNTIWGACFAKSAACYNPIVYGISH PKYRLALKEKCPCCVFGKVDDGKSSEAQSQATTSEAESKA >INSE_MWS_calEry Calliphora erythrocephala M58334 490 Arthropoda Insecta Rh1 complete MERYSTPLIGPSFAALTNGSVTDKVTPDMAHLVHPYWNQFPAME PKWAKFLAAYMVLIATISWCGNGVVIYIFSTTKSLRTPANLLVINLAISDFGIMITNT PMMGINLFYETWVLGPLMCDIYGGLGSAFGCSSILSMCMISLDRYNVIVKGMAGQPMT IKLAIMKIALIWFMASIWTLAPVFGWSRYVPEGNLTSCGIDYLERDWNPRSYLIFYSI FVYYLPLFLICYSYWFIIAAVSAHEKAMREQAKKMNVKSLRSSEDADKSAEGKLAKVA LVTISLWFMAWTPYTIINTLGLFKYEGLTPLNTIWGACFAKSAACYNPIVYGISHPKY GIALKEKCPCCVFGKVDDGKASDATSQATNNESETKA >INSE_MWS_droMel Drosophila melanogaster M12896 420 Arthropoda Insecta Rh2 complete genNow MERSHLPETPFDLAHSGPRFQAQSSGNGSVLDNVLPDMAHLVNP YWSRFAPMDPMMSKILGLFTLAIMIISCCGNGVVVYIFGGTKSLRTPANLLVLNLAFS DFCMMASQSPVMIINFYYETWVLGPLWCDIYAGCGSLFGCVSIWSMCMIAFDRYNVIV KGINGTPMTIKTSIMKILFIWMMAVFWTVMPLIGWSAYVPEGNLTACSIDYMTRMWNP RSYLITYSLFVYYTPLFLICYSYWFIIAAVAAHEKAMREQAKKMNVKSLRSSEDCDKS AEGKLAKVALTTISLWFMAWTPYLVICYFGLFKIDGLTPLTTIWGATFAKTSAVYNPI VYGISHPKYRIVLKEKCPMCVFGNTDEPKPDAPASDTETTSEADSKA >CRUS_LWS_meoOer Neogonodactylus oerstedii DQ646869 489 Arthropoda Crustacea Rh1 complete MSYWNSNKIVEEYSLPSTNPYGNFTVVDTVPENMLHMIHSHWYQ FPPLNPMWYGILAFVVTVVGLCSICGNFVVIWVFMNTKALRSPANTLVVSLAVSDFIM MACMFPPLVLNCYWGTWIFGPLFCEVYAFIGNTVGCASIGNMIFITFDRYNVIVKGIS GTPLSQKNTTLQVLFVWICSIMWCVFPFFGWNRYVPRGDMTACGTDYLTEDEFSRSYL YVYSVWVYIGPLALIIYCYFHIVSAVATHEKQMRDQAKKMGVKSLRTEEAKKTSAECR LAKVALTTVSLWFMAWTPYLIINWAGMFYPSVVSPLFSIWGSVFAKANAVYNPIVYAI SHPKYRAALYKKLPCLACSTESADEGSATNSATTTTAEKYESA >CRUS_LWS_neoOer Neogonodactylus oerstedii DQ646871 522 Arthropoda Crustacea Rh3 complete MSYWNSNKAMEEYSLPSTNPYGNFTVVDTVPENMLHMVHSHWYQ FPPLNPMWYGILAFVVTVVGLCSICGNFVVIWVIMNTKALRSPANTLVVSLAVSDYIM MTCMFPPLVLNCYWGTWIFGPLFCEVYAFIGNTVGCASIGNMIFITFDRYNVIVKGVS GKPLSQKNATLQVLFVWICSIMWCVFPFFGWNRYVPEGNMTACGTDYLTEDEFSRSYL YIYSVWVYIGPLALIIYCYFHIVSAVATHEKQMRDQAKKMGVKSLRTEEAKKTSAGCR LAKVALTTVSLWFMAWTPYLIINWAGMFYPSVVSPLFSIWGSVFAKSNAVYNPIVYAI SHPKYRAALYKKLPCLACSTESADEGSATNSTTTATAEKYESA >CRUS_LWS_eupSub Euphausia superba DQ852576 487 Arthropoda Crustacea DQ852580 partial MNPLWYGLLGFVIFCLGCLSVFGNSVVIWVFTSTKTLRSPANML VVNLALSDFLMMANMSPPTVHSCYHGTWMLGPTYCEYYALVGSLSGCISIWTMVWITL DRYNVIVKGVAATPLTNKGAFARNIFSWLSALIWCVSPLYGWNRYVPEGNMTACGTDY LTDDWLSHSYLYAYTFWVYLFPFFIIVYCYTYIVSAVFAHEKGMRDQAKKMGVKSLRN EEAQKTSAECRLAKVALVTVSLWFIAWTPYCVINVTGMWDKTKITPLFTIWGSL >CRUS_LWS_homGam Homarus gammarus DQ852587 515 Arthropoda Crustacea DQ852590 partial MNPLWYGLLALWMFVMGTLSVCGNSIVIWVFMNTKALRTPANLL VVNLAISDFLMMFCMCPPLLINCYYQTWVWGAFACEVYGCIGSTVGTCSIFCMVFITM DRYNVIVKGVSATPLTTNGAMLRNLFSWVTSIGWCLPPFFGFNAYVPEGNLIACGTDY LKESVPYHVYLYLYSVWCYFLPLVIIVYCYTYIVAAVSAHERQMREQAKKMGVKSLRS EESKKTSNECRLAKVALTTVSLWFIAWTPYLIINWAGMINKPSVSPLLTI >CRUS_LWS_camLud Cambarus ludovicianus AF003543 529 Arthropoda Crustaceapartial LHMIHLHWYQYPPMNPMMYPLLLVFMLITGILCLAGNFVTIWVF MNTKSLRTPANLLVVNLAMSDFLMMFTMFPPMMITCYYHTWTLGATFCEVYAFLGNLC GCASIWTMVFITFDRYNVIVKGVAGEPLSTKKASLWILTVWVLSFTWCVAPFFGWNRY VPEGNLTGCGTDYLSEDILSRSYLYIYSTWVYFLPLAITIYCYVFIIKAVAAHEKGMR DQAKKMGIKSLRNEEAQKTSAECRLAKIAMTTVALWFIAWTPYLLINWVGMFARSYLS PVYTIWGYVFAKANAVYNPIVYAIS >CRUS_LWS_proMil Procambarus milleri AF003546 522 Arthropoda Crustaceapartial LHMIHLHWYQYPPMNPMMYPLLLIFMLFTGILCLAGNFVTIWVF MNTKSLRTPANLLVVNLAMSDFLMMFTMFPPMMVTCYYHTWTLGPTFCQVYGFLGNLC GCASIWTMVFITFDRYNVIVKGVAGEPLSTKKASLWILIVWVLSLAWCMAPFFGWNRY VPEGNLTGCGTDYLSEDILSRSYLYIYSTWVYFLPLTITIYCYVFIIKAVAAHEKGMR DQAKKMGIKSLRNEEAQKTSAECRLAKIAMTTVALWFIAWTPYLLINWVGMFARSYLS PVYTIWGYVFAKANAVYNPIVYAIS >CRUS_LWS_arcGre Archaeomysis grebnitzkii DQ852573 496 Arthropoda Crustacea DQ852575 partial MNPLWYGLLGFVIFCLGILSVCGNAVVIWVFMNTKSLRSPANLL VVNLAFSDFLMMLNMFPPMVHSCYHGTWMLGAFFCEFYGFTGSLFGCISIWTMVFITM DRYNVIVKGVAAEPLTSKGASIRILFVWTVAFAWTILPFFGWNRYVPEGNLTACGTDY LTEDSTSHLYLYMYASWAYYTPLLYIIYAYTFIVQAVSAHEKGMREQAKKMGVKSLRN EEAQKTSAECRLAKVALMTVSLWFMAWTPYMIINFTGMNDRTKLTPLCTIWGSL >CRUS_LWS_holCos Holmesimysis costata DQ852581 512 Arthropoda Crustacea DQ852586 partial MNPLWYGLLGFWMTVMGTLSVAGNFVVIWVFMNTKSLRTPANLL VVNLAISDFFMMLTMTPPLLANAYWGTWILGAFFCEVYAFLGSFFGCVSIWSMVFITA DRYNVIVKGVSAEPLTSGGAMMRIAGTWAFTLAWCLPPFFGWNRYVPEGNMLACGTDY LTETELSRSYLYVYSVWVYLFPLAYIIYSYTFIVKAVAAHEKGMREQAKKMGVKSLRS EEAQKTSAECRLCKVALMTVTLWFMAWTPYFIINWGGMFNKPMVTPLFS >CRUS_LWS_mysDil Mysis diluviana DQ852591 501 Arthropoda Crustaceapartial MKSRWYIILGLIISVLAILSVIGNLTVIVVFINTRSLRSPSNLL IVNLAFSDFFMMCNMCPAMLLACIYKTWLLGPTYCAWYAFSGSLFGCLSIWTMVWITL ERYNVIVKGVSSKPLSVKGAITRIVLTWIFAVIWCSFPLVGWNRYVPEGNLTACGTDY LSDDIYSQSYIYLYSVMVYFIPLGITIYCYSYIVHAVANHEKSMKEQAKKMGVKSFRN EETQRTSAEFRLAKIALMTVSLWFIAWTPYLVINIVGMVARQQLNPLSTI >CRUS_LWS_neoAme Neomysis americana DQ852592 520 Arthropoda Crustacea DQ852598 partial MNPLWYSLVGFWMVIMGVLSVVGNFVVLWVFMTTKSLRTPANLL VVNLALSDFLMMFTMFPPMVISCYWQTWTLGAFFCEVYAFLGSLFGCVSIWSMVWITL DRYNVIVKGVSGEPLTNSGAMTRIAGTWVTAFAWCLPPFFGWNRYVPEGNMTACGTDY LTDDKFSHSYLYIYSVWVYIFPLFLNIYLYTFIIKAVANHEKQMREQAKKMGVKSLRS EESQKTSAECRLAKVALMTVSLWFMAWTPYFIINWAGMLSKSNVTPLFSIWGSV >CHEL_LWS_limPol Limulus polyphemus L03782 530 Arthropoda Chelicerata ocelli complete genFut MANQLSYSSLGWPYQPNASVVDTMPKEMLYMIHEHWYAFPPMNP LWYSILGVAMIILGIICVLGNGMVIYLMMTTKSLRTPTNLLVVNLAFSDFCMMAFMMP TMASNCFAETWILGPFMCEVYGMAGSLFGCASIWSMVMITLDRYNVIVRGMAAAPLTH KKATLLLLFVWIWSGGWTILPFFGWSRYVPEGNLTSCTVDYLTKDWSSASYVIIYGLA VYFLPLITMIYCYFFIVHAVAEHEKQLREQAKKMNVASLRANADQQKQSAECRLAKVA MMTVGLWFMAWTPYLIIAWAGVFSSGTRLTPLATIWGSVFAKANSCYNPIVYGISHPR YKAALYQRFPSLACGSGESGSDVKSEASATMTMEEKPKSPEA >CRUS_MWS_hemSan Hemigrapsus sanguineus D50583 480 Arthropoda Crustacea D50584 complete MANVTGPQMAFYGSGAATFGYPEGMTVADFVPDRVKHMVLDHWY NYPPVNPMWHYLLGVVYLFLGVISIAGNGLVIYLYMKSQALKTPANMLIVNLALSDLI MLTTNFPPFCYNCFSGGRWMFSGTYCEIYAALGAITGVCSIWTLCMISFDRYNIICNG FNGPKLTQGKATFMCGLAWVISVGWSLPPFFGWGSYTLEGILDSCSYDYFTRDMNTIT YNICIFIFDFFLPASVIVFSYVFIVKAIFAHEAAMRAQAKKMNVTNLRSNEAETQRAE IRIAKTALVNVSLWFICWTPYAAITIQGLLGNAEGITPLLTTLPALLAKSCSCYNPFV YAISHPKFRLAITQHLPWFCVHEKDPNDVEENQSSNTQTQEKS >INSE_UVV_camAbd Camponotus abdominalis AF042788 360 Arthropoda Insecta complete MYNGSFHWEARILPAGPPRLLGWNVPAEELVHIPEHXLVYPEPN PSLHYLLAIVYILFTFVALFGNGLVIWIFCSAKSLRTPSNLFVVNLAFCDFMMMLKAP IFIYNSFHTGFATGHLGCQIFACMGSLSGIGAGMTNAAIAYDRYSTIARPLDGKLSRG QVLLLIMLIWTYTIPWALMPLMQVWGRFVPEGFLTSCSFDYLTDSQEIRYFVPTIFTF SYCVPMLLIIYYYSQIVGHVVSHEKALREQAKKMNVESLRSNVNTNAQSAEIRIAKAA ITICFLFVLSWTPYGALAMIGAFGNRALLTPGITMIPACACKFVACLDPYVYAISHPR YRLELQKRLPWLELQEKPVADTQSTTTEMVHTPAS >INSE_UVV_catBom Cataglyphis bombycinus AF042787 360 Arthropoda Insecta complete MYTNRSVHWEARILPAGPPRLLGWNVPAEELVHIPEHWLVYPEP NPSLHYLLAILYTLFTFVALLGNGLVIWIFISAKSLRTPSNMFVVNLAFCDFIMMLKA PIFIYNSFNTGFATGHLGCQIFACMGALSGIGASMTNAAIAYDRYSTIARPLDGKLSR GQVILLIALIWTYTIPWALMPLMHVWGRFVPEGFLTSCTFDYLTDTPEIRYFVATIFT FSYCIPMSLIIYYYSQIVSHVVNHEKALREQAKKMNVESLRSNTNTNAQSAEIRIAKA AITICFLFVLSWTPYGTLAMIGAFGNKALLTPGVTMIPACTCKFVACLDPYVYAISHP RYRLELQKRLPWLELQEKPIETQSTTTETVNTASS >INSE_UVV_manSex Manduca sexta L78081 357 Arthropoda Insecta complete genFut MNNQSENYYHGAQFEALKSAGAIEMLGDGLTGDDLAAIPEHWLS YPAPPASAHTALALLYIFFTFAALVGNGMVIFIFSTTKSLRTSSNFLVLNLAILDFIM MAKAPIFIYNSAMRGFAVGTVGCQIFALMGAYSGIGAGMTNACIAYDRHSTITRPLDG RLSEGKVLLMVAFVWIYSTPWALLPLLKIWGRYVPEGYLTSCSFDYLTNTFDTKLFVA CIFTCSYVFPMSLIIYFYSGIVKQVFAHEAALREQAKKMNVESLRANQGGSSESAEIR IAKAALTVCFLFVASWTPYGVMALIGAFGNQQLLTPGVTMIPAVACKAVACISPWVYA IRHPMYRQELQRRMPWLQIDEPDDTVSTATSNTTNSAPPAATA >INSE_UVV_papXut Papilio xuthus AB028218 --- Arthropoda Insecta Rh5 partial MIPAAVMDNHTENNYNYGAYFAPYRLEGVELLGAGLTGEDLAAI PEHWLSYPAPPASAHTMLALVYVFFTAAALIGNGLVIFIFSASKSLRTPSNLLVVQLA VLDFLMMLKAPIFIYNSIKRGFASGVIGCQIFAFMGSVSGTAAGLTNACIAYDRHSTI TRPLDGRLSRGKVLLMMVCVWLYTAPWAILPQLQIWGRYVPEGFLTSCTFDYLTTTFD NKLFVASMFVCVYIFPMIAILYFYSGIVKQVFAHEAALREQAKKMNVDSLRSNQNAAA ESAEIRIAKAALTVCFLYVASWTPYGVMSLIGAFGDQNLLTPGVTMIPALACKGVACI DPWVYAISHPKYRQELQKRMPWLQIDEPDDNASNTTSNTANSSAPA >INSE_UVV_droMel Drosophila melanogaster NM_057353 375 Arthropoda Insecta Rh4 complete genNow MEPLCNASEPPLRPEARSSGNGDLQFLGWNVPPDQIQYIPEHWL TQLEPPASMHYMLGVFYIFLFCASTVGNGMVIWIFSTSKSLRTPSNMFVLNLAVFDLI MCLKAPIFIYNSFHRGFALGNTWCQIFASIGSYSGIGAGMTNAAIGYDRYNVITKPMN RNMTFTKAVIMNIIIWLYCTPWVVLPLTQFWDRFVPEGYLTSCSFDYLSDNFDTRLFV GTIFFFSFVCPTLMILYYYSQIVGHVFSHEKALREQAKKMNVESLRSNVDKSKETAEI RIAKAAITICFLFFVSWTPYGVMSLIGAFGDKSLLTPGATMIPACTCKLVACIDPFVY AISHPRYRLELQKRCPWLGVNEKSGEISSAQSTTTQEQQQTTAA >INSE_UVV_droMel Drosophila melanogaster M17718 345 Arthropoda Insecta Rh3 complete MESGNVSSSLFGNVSTALRPEARLSAETRLLGWNVPPEELRHIP EHWLTYPEPPESMNYLLGTLYIFFTLMSMLGNGLVIWVFSAAKSLRTPSNILVINLAF CDFMMMVKTPIFIYNSFHQGYALGHLGCQIFGIIGSYTGIAAGATNAFIAYDRFNVIT RPMEGKMTHGKAIAMIIFIYMYATPWVVACYTETWGRFVPEGYLTSCTFDYLTDNFDT RLFVACIFFFSFVCPTTMITYYYSQIVGHVFSHEKALRDQAKKMNVESLRSNVDKNKE TAEIRIAKAAITICFLFFCSWTPYGVMSLIGAFGDKTLLTPGATMIPACACKMVACID PFVYAISHPRYRMELQKRCPWLALNEKAPESSAVASTSTTQEPQQTTAA >INSE_BLU_manSex Manduca sexta AD001674 450 Arthropoda Insecta complete genFut MATNFTQELYEIGPMAYPLKMISKDVAEHMLGWNIPEEHQDLVH DHWRNFPAVSKYWHYVLALIYTMLMVTSLTGNGIVIWIFSTSKSLRSASNMFVINLAV FDLMMMLEMPLLIMNSFYQRLVGYQLGCDVYAVLGSLSGIGGAITNAVIAFDRYKTIS SPLDGRINTVQAGLLIAFTWFWALPFTILPAFRIWGRFVPEGFLTTCSFDYFTEDQDT EVFVACIFVWSYCIPMALICYFYSQLFGAVRLHERMLQEQAKKMNVKSLASNKEDNSR SVEIRIAKVAFTIFFLFICAWTPYAFVTMTGAFGDRTLLTPIATMIPAVCCKVVSCID PWVYAINHPRYRAELQKRLPWMGVREQDPDAVSTTTSVATAGFQPPAAEA >INSE_BLU_apiMel Apis mellifera AF004168 439 Arthropoda Insecta complete genNow MLLHNKTLAGKALAFIAEEGYVPSMREKFLGWNVPPEYSDLVHP HWRAFPAPGKHFHIGLAIIYSMLLIMSLVGNCCVIWIFSTSKSLRTPSNMFIVSLAIF DIIMAFEMPMLVISSFMERMIGWEIGCDVYSVFGSISGMGQAMTNAAIAFDRYRTISC PIDGRLNSKQAAVIIAFTWFWVTPFTVLPLLKVWGRYTTEGFLTTCSFDFLTDDEDTK VFVTCIFIWAYVIPLIFIILFYSRLLSSIRNHEKMLREQAKKMNVKSLVSNQDKERSA EVRIAKVAFTIFFLFLLAWTPYATVALIGVYGNRELLTPVSTMLPAVFAKTVSCIDPW IYAINHPRYRQELQKRCKWMGIHEPETTSDATSAQTEKIKTDE >INSE_BLU_droMel Drosophila melanogaster U67905 437 Arthropoda Insecta Rh5 complete genNow MHINGPSGPQAYVNDSLGDGSVFPMGHGYPAEYQHMVHAHWRGF REAPIYYHAGFYIAFIVLMLSSIFGNGLVIWIFSTSKSLRTPSNLLILNLAIFDLFMC TNMPHYLINATVGYIVGGDLGCDIYALNGGISGMGASITNAFIAFDRYKTISNPIDGR LSYGQIVLLILFTWLWATPFSVLPLFQIWGRYQPEGFLTTCSFDYLTNTDENRLFVRT IFVWSYVIPMTMILVSYYKLFTHVRVHEKMLAEQAKKMNVKSLSANANADNMSVELRI AKAALIIYMLFILAWTPYSVVALIGCFGEQQLITPFVSMLPCLACKSVSCLDPWVYAT SHPKYRLELERRLPWLGIREKHATSGTSGGQESVASVSGDTLALSVQN >INSE_UVV_apiMel Apis mellifera AF004169 353 Arthropoda Insecta complete genNow MSNDSIHWEARYLPAGPPRLLGWNVPAEELIHIPEHWLVYPEPN PSLHYLLALLYILFTFLALLGNGLVIWIFCAAKSLRTPSNMFVVNLAICDFFMMIKTP IFIYNSFNTGFALGNLGCQIFAVIGSLTGIGAAITNAAIAYDRYSTIARPLDGKLSRG QVILFIVLIWTYTIPWALMPVMGVWGRFVPEGFLTSCSFDYLTDTNEIRIFVATIFTF SYCIPMILIIYYYSQIVSHVVNHEKALREQAKKMNVDSLRSNANTSSQSAEIRIAKAA ITICFLYVLSWTPYGVMSMIGAFGNKALLTPGVTMIPACTCKAVACLDPYVYAISHPK YRLELQKRLPWLELQEKPISDSTSTTTETVNTPPASS