Opsin evolution: key critters (protostomes): Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 461: | Line 461: | ||
Every aspect of photoreception in Drosophila has been studied for decades. Because this research is [http://mbe.oxfordjournals.org/cgi/content/full/26/2/299#BIB57 regularly reviewed] at length, the focus here is on genome project developments and issues that remain in characterizing opsin function and evolution. | Every aspect of photoreception in Drosophila has been studied for decades. Because this research is [http://mbe.oxfordjournals.org/cgi/content/full/26/2/299#BIB57 regularly reviewed] at length, the focus here is on genome project developments and issues that remain in characterizing opsin function and evolution. | ||
Drosophila has seven opsins, all of melanopsin class. Ciliary-class opsins (present elsewhere in arthropods) have been lost, as have the peropsins classes (which persist in deuterostomes and some lophotrochozoa perhaps because without ciliary opsins there is no need for a retinal isomerase regeneration cycle). The paired Drosophila retinas have 850 ommatidia each housing eight photoreceptors of three types; the paired cephalopharyngeal Bolwig organs have 12 photoreceptor cells of two types. Oddly, during metamorphosis to eyelet, the outer Bolwig cells die while inner cells [http://www.ncbi.nlm.nih.gov/pubmed/18594514 switch gene expression] from Rh6 to RH5. | Drosophila has seven opsins, all of melanopsin class. Ciliary-class opsins (present elsewhere in arthropods) have been lost in all 12 drosophilid genomes, as have the peropsins classes (which persist in deuterostomes and some lophotrochozoa perhaps because without ciliary opsins there is no need for a retinal isomerase regeneration cycle). However it raises the question whether some neuroanatomical structures have also been lost. | ||
The paired Drosophila retinas have 850 ommatidia each housing eight photoreceptors of three types; the paired cephalopharyngeal Bolwig organs have 12 photoreceptor cells of two types. Oddly, during metamorphosis to eyelet, the outer Bolwig cells die while inner cells [http://www.ncbi.nlm.nih.gov/pubmed/18594514 switch gene expression] from Rh6 to RH5. | |||
Two of the Drosophila opsins have peak sensitivity in the ultraviolet (RH5 RH7) consistent with their K90 lysine and shorter motif, two sister opsins peak (RH3 RH4) in the blue and the rest (R6,(RH1,RH2)) at longer visible wavelengths. Opsins have been assigned to the four known photoreceptor structures as follows: | Two of the Drosophila opsins have peak sensitivity in the ultraviolet (RH5 RH7) consistent with their K90 lysine and shorter motif, two sister opsins peak (RH3 RH4) in the blue and the rest (R6,(RH1,RH2)) at longer visible wavelengths. Opsins have been assigned to the four known photoreceptor structures as follows: | ||
Revision as of 16:39, 18 June 2009
Key Critters: introduction to genome projects opsins
Some species such as Drosophila have lost all ciliary opsins -- clearly this class of genes is not essential for a successful visually complex flying insect with 5-color vision, periferal motion detection, polarized light capability and circadian rhythm (as one might have assumed from vertebrates). Other protostome lineages such as nematodea (eg Caenorhabditis elegans) function successfully without any vision at all, making this 'model organism' completely irrelevent to the evolutionary study of vision.
However bees, annelids, and mammals retain ciliary opsins so it follows -- pervasive, detailed convergence at the molecular level being impossible -- this must be the ancestral bilateran state state. In turn that suggests ciliary opsins in cnidaria and indeed that has been recently established in the lensing eye.
When the eye is reduced to a single pigment cell backing a single photoreceptor cell, the opsin of that species may be expressed only in one cell of the entire body. In this situation, the opsin may never show up in transcript collections, even with subtraction of common ones. One sees the importance of complete genomes here (versus transcripts or immunostained sections alone): absence of ciliary opsin evidence in a genome is truly evidence of ciliary opsin absence.
Vertebrates could never have evolved ciliary opsin vision had the bilateran ancestor possessed the limited opsin repertoire of fruit fly. Thus the most pressing question is -- assuming rhabdomeric opsins were thoroughly entrenched in the earliest bilateran imaging eyes and photoreception systems -- what kept ciliary opsins around in early bilatera? Recall early diverging deuterostomes (xenoturbellids, urchins, acorn worms, tunicates, and lancelets) lack imaging vision -- that emerged in full modern form on the lamprey stem.
Conversely, assuming cnidaria use ciliary opsins, what kept rhabdomeric opsins around so that they could later be co-opted by protostomes for their form of opsin-based vision? Evolution is strictly 'use it or lose it' over these time frames. Here cnidaria, or at least their larva, may also use rhabdomeric opsins. It seems that both classes of opsins have retained roles in most species, but very different classes were promoted to the imaging role in different branches of Bilatera. In fly, ciliary opsins have winked out; in nematode, both ciliary and rhabdomeric opsins are gone. While irrevocable, these losses would scarcely receive comment in non-model organisms.
It's important to understand contemporary representatives of early diverging species (relative to the sequence of divergence nodes leading to human) are not archaic failed experiments nor primitive living fossils frozen in evolutionary time. Quite the contrary, all surviving extant species are equally successful and fully modern -- the tree of life is right-justified. Indeed their genes, regulatory signalling systems, and enzymes may be more finely honed than slowly evolving mammals because of more rapid evolution attributable to larger effective population sizes, reproductive mode, short generation time, and marine selective predatory pressures.
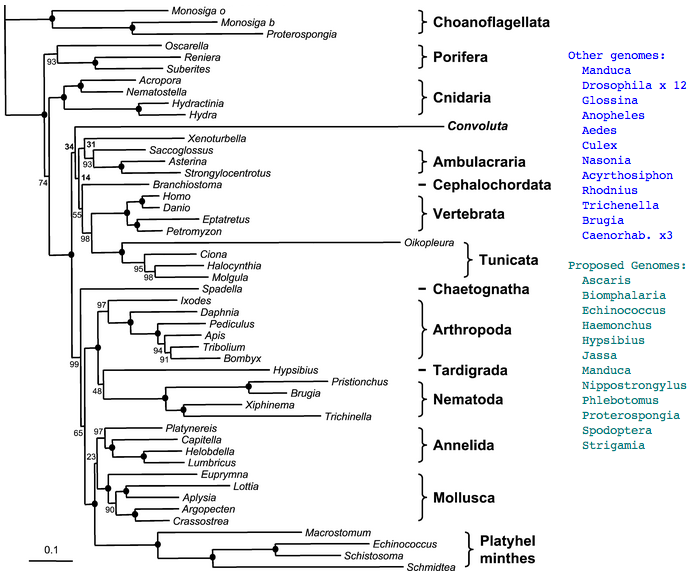
However we can still hope that ancestral character traits will still be reflected to some extent in these earlier diverging species and that with enough complete opsin repertoires from taxonomically appropriate species, ancestral genes and even whole visual systems can be reconstructed at key ancestral nodes on the phylogenetic tree. The story describing the evolution of the human eye then amounts to describing its status at these successive nodes with perhaps interpolative speculation between them. Definitely limits to knowledge exist because living metazoans provide only 35 nodes between sponge and human -- gaps between nodes may average 30 million years but can greatly exceed that (eg 135 myr between bird and platypus). This is offset by the occasional proposal for new deuterostome branches (Xenoturbella, Convoluta) or basal metazoan (Ctenophores.
The ideal set of genomes needed to study the evolution of the metazoan eye is only partly completed, underway, or not even proposed yet. In some cases, the genome size of clade representatives is so large (eg lungfish at 25x human) the species may never be sequenced, though satisfactory opsin transcripts could still be obtained. In others, the rate of evolution has been so fast so long that very little information about photoreception at ancestral nodes has been retained (eg the tunicate Oikopleura). Hagfish opsins, which would conveniently break up the crucial lamprey long branch, are not available at GenBank but here the animal has adopted a deep water habitat, meaning that its cone opsin genomic repertoire will be highly reduced, if not gone entirely, in its markedly degenerated eyes, though whatever remains of its opsins could still be informative.
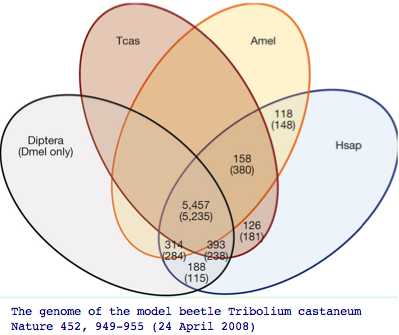
The impact of adding more genomes is to uncover more genes of the common bilateran ancestor that were masked by lineage-specific losses. Recall the beatle genome Tribolium uncovered 126 additional genes absent in other insect genomes but nonetheless present in human. Humans themselves of course have lost hundreds of genes even relative to the first land animal, so here too we need to pool mammalian and amniote gene pools to reconstruct that ancestor.
Model organism choices do not always coincide with genome sequenceability, transcriptome projects, nor (worst of all) with more slow-evolving and less derived species. Finally, most sequencing speaks to narrow anthropocentric interests, whereas the sequencing need more broadly conceived is greatest farther back (to break up long branches). The evolution of the eye needs a rather different portfolio of genomes than a typical human disease gene because of the earlier intrinsic timing of the innovative events. In fact, one product of the investigation here is to spell out these needed genomes. Of course one obvious genome choice are cubomedusan jellyfish with their 24 eyes of 6 types.
It's worth reviewing genome status and recent experimental literature on key species. While abstracts are readily available at PubMed, access to free full text is unpredictable, so those links are collected when available. It suffices to reference only recent articles because those in turn cite the earlier literature and citation in turn of their paper are collected by Google Scholar (or AbstractsPlus at PubMed). Most opsin sequences in the Opsin evolution reference sequence collection have a PubMed accession as a field in their fasta header database; those can simply be compiled to an active link that opens all of them in one PubMed window.
Figure adapted from: Acoel Flatworms Are Not Platyhelminthes: Evidence from Phylogenomics (H Philippe et al PLoS ONE. 2007 Aug 8)
Deuterostomes moved to separate article
The key critter article has been broken down into 3 smaller articles -- deuterostomes are now here.
Chondrichthyes: Callorhinchus milii (elephantshark) 13 opsins Agnatha: Petromyzon marinus (lamprey) 9 opsins Agnatha: Eptatretus burgeri (hagfish) 0 opsins Urochordata: Ciona intestinalis (tunicate) 4 opsins Echinodermata: Stronglyocentrotus purpuratus (sea urchin) 6 opsins Hemichordata: Saccoglossus kowalevskii (acornworm) 1 opsin Deuterostomia: Xenoturbella bocki + Convoluta pulchra 0 opsins
Cnidaria and Porifera moved to separate article
The key critter article has gotten too large -- cnidaria are now here.
Cubozoa: Tripedalia cystophora .. 1 ciliary opsin Cubozoa: Carybdea marsupialis (jellyfish) .. probable opsins Anthozoa: Nematostella vectensis (sea anemone) .. claimed opsins Hydrozoa: Hydra magnipapillata (hydra) .. claimed opsins Hydrozoa: Cladonema radiatum (jellyfish) .. claimed opsins Porifera, Placozoa, Choanoflagellates .. 0 opsins
Lophotrochozoa: 13 opsins
This is a monophyletic group (in the mind of evo-devo practitioners) of bilaterans reflecting a [basal split deep within protostomes. The classification is based both on molecular considerations and a shared larval form with ciliated wheel, in contrast to characters of adult animals such as segmentation.
Lophotrochozoa is not recognized at GenBank so blast searches cannot be restricted to Lophotrochozoa. However Entrez and PubMed searches can be so restricted using boolean queries. In terms of genome projects Lophotrochozoa currently consists of 7 species of flatworms, molluscs, and annelids. However, it also contains Brachiopoda, Bryozoa, Entoprocta, Nemertea, Sipuncula, etc which collectively account for less than 3,000 of the 5.7 million nucleotide sequences at GenBank and no annotated opsins.
The Lophotrochozoa have not been surveyed as a whole for those that might be 'living fossils' in terms of opsins and photoreceptor structures. Even those would not necessarily make good genome projects because of genome size and compositional issues. However Annelida has been thoroughly considered by Purschke, Arendt et al in a recent offline, off-Pubmed review (Arthropod Structure & Development 35(2006) 211-230).
Annelida: Platynereis dumerilii (ragworm) .. 3 opsins
This small annelid may be an emerging model organism, though plans for genome sequencing in France have apparently collapsed. (Indeed all metazoan genomic sequencing in Europe has ceased.) Three recent papers have established that Platynereis qualifies as living fossil, at least with respect to ancestral anatomy and development, slowly evolving protein sequences and retention of genes and ancestral introns, and further has retained ciliary opsins.
That is to say, fruit fly and nematode have proven unfortunate choices because of so many lost genes and signalling pathways, rapid evolution and highly derived characters. Lophotrochozoa may thus give us very significant insight into the bilateran ancestor that had appeared lost from consideration of just Arthropoda. It should be noted though that not all insects should be written off, Anopheles has also retained ciliary opsins.
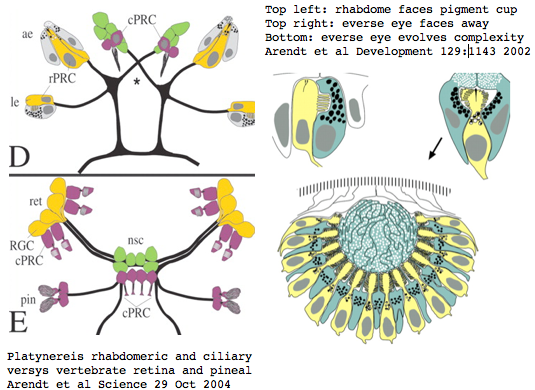
Platynereis develops various pairs of eyes going by localization of opsin expression: inverse larval eyes used in phototaxis (just one pigment cell and one photoreceptor cell) and two pairs of everse adult eyes needed for adult vision. These originate from an initially unsplit single anlage. These eyes use exclusively rhabdomeric photoreceptor cells and corresponding rhabdomeric-class opsins as expected from phylogenetic position. However two paired structures in the developing median brain dorsal to the apical organ express an opsin that unambiguously classifies as ciliary. Further, a retinal homeobox (specific to ciliary pineal eyes) and circadian rhythm regulator bmal are also expressed at this location in Platynereis. However the pigment cells necessary for directional photoreception are missing. This all fits with a role for the ciliary opsin as the primary receptor underlying circadian rhythm which does not require directionality.
The emerging picture is Urbilatera having both ciliary and rhabdomeric structures. The later specialized structure was lost but the photoreceptor component retained in vertebrates in the form of melanopsins expressed in retinal ganglion cells.
Remarkably, Platynereis contains a second ciliary opsin next to alpha tubulin: Using the initial ciliary opsin (a transcript with unknown intronation) as probe at various GenBank databases, a genomeWiki contributer found a 171,779 bp survey sequence in the high throughput genomic sequence HTGS division (meaning it would be overlooked using Blast of the nucleotide division) had a good match in the unannotated contig CT030681, submitted 05-DEC-2005 by Genoscope as 6 ordered contigs (the last of which proves reverse-complemented).
This second opsin, being genomic, after difficult recovery of full length gene from a moderate match, could be intronated (unlike the original transcript) assuming GT-AG splice junctions (like 99% of all genes and 100% of all known opsins). These introns had positions and phases identical to ciliary -- but not Go or Gq -- deuterostome opsins. Assuming the first opsin is not derived as a processed retrogene from the first, it can be intronated via homological alignment. These are stored in the Opsin Classifier as CILI1_plaDum and CILI2_plaDum, resp.
Using the second opsin as blastp query against our phylogenetically dispersed collection of 225 hand-curated Eumetazoan opsins (including new cnidarian ciliary opsins), it classifies in the encephalopsin-to-pinopsin area in accord with independent classification by intron pattern and close homology with the experimentally characterized Platynereis first opsin. The percent identity to deuterostome opsins is not only quite high (considering the immense round-trip time since common ancestor) but also overwelmingly concentrated on invariant and near-invariant amino acids characteristic of ciliary opsins. Thus this second Platynereis opsin cannot be a pseudogene (unless that happened yesterday or so).
For purposes of conserved synteny [eg establishing orthology to related opsins in other lochotrophoran genomes], other coding genes on this contig using blastx vs metazoan proteins) can be considered. The only other gene is alpha-tubulin, at positions 124517-122811, downstream from the second ciliary opsin at 46848-87956 using original contig ordering.
Recall the Arendt group used antibody to acetylated alpha-tubulin was used as marker for stabilized microtubules in cilia and axons. They needed the sequence for that. Probably the larger contig was then sequenced as part of the genome feasibility survey. There was no particular reason to look at this contigs for opsins at that time, which would be hard to distinguish from abundant non-photoreceptor rhodopsin-superfamily genes or generic GPCR.
Supposing Platynereis has 15,000 coding genes, this is quite a coincidence to have two genes adjacent that might be critical to the same photoreceptor structure. If these two genes are transcribed divergently (lie on different strands) after fixing (reverse-complementing) the last contig piece, then symmetric transcriptional regulatory element DNA (read the same whichever strand), this could mean the second opsin is tethered to alpha-tubulin production in terms of co-expression in some cell types. Transcribed in the same direction is less attractive as operons are rare in eukaryotes, though read-through is not unheard of and that too could be developmentally regulated in extent.
Re-assembly CT030681 using multi-exon bridging is possible. It turned out pieces 1 and 2 were irrelevent, piece 3 had exons 1,2,3 of the opsin on the plus strand, piece 4 had opsin exon 4 and 5 on the minus strand to piece-coordinate 41,899 for the stop codon. This piece also contains the first three exons of alpha tubulin also on the minus strand beginning at 36,767. Its initial methionine is stranded as a solitary phase 0 codon on the end of 5' UTR, 36,707-05. The remaining two exons of alpha tubulin are on the minus strand of piece 5.
Joining piece 3 with reverse-complemented pieces 4 and 5 then fixes orientations to the plus strand and establishes intron sizes subject to the two strings of Ns. This results in parallel gene order CILI2_plaDum+ TUBA_plaDum+, that is tubilin downstream of the opsin with an intergenic gap of 5,132 bp. If there is any coordination of expression by read-thru, on the upstream end it would have to involve the regulatory regions of the opsin.
The fifth exon of CILI2_plaDum has too weak match with that of CILI1_plaDum to be found by conventional searching. However the dna where it has to be located is squeezed between exon 4 and the start of tubulin, reducing query size. Blastx of that dna against the full-blown set of opsins turns up a consistent match candidate in frog and skate opsins. Looking at the intron phasing validates the match since the splice acceptor AG is 1 of 16 dinucleotides, the phase 0 required by exon 4 (and ancestral ciliary phase) is 1 of 3 possible phases, and 1 of 2 strand requirement have together a 1 in 96 chance of random occurence, more than sufficient in conjunction with the blast expectation of 1.1e-06.
This opsin if co-expressed with CILI1_plaDum would amount to 'circadian rhythm color vision'. Alternately it might be expressed at a different developmental stage or in an unsuspected auxillary photoreceptor.
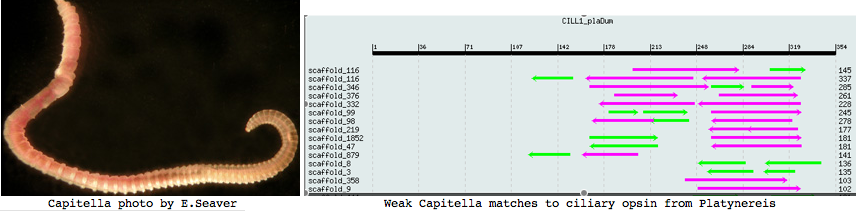
Annelida: Capitella sp (marine worm) .. 2 opsins
Capitella is a small segmented benthic marine worm most closely related, in the genome project sense, to its fellow annelid Platynereis. The taxonomy of the genus Capitella was thoroughly muddled by a quaint 1976 starch gel electrophoresis allozyme study; Linnean nomenclature has never been developed for the 6 alleged species defined there. The isolate used in the JGI genome project is called Capitella sp. I ES-2005 instead of Capitella capitata.
The last of 3,709,316 trace reads were taken in Nov 2005. As with Lottia, a multi-year lag ensued in release of the assembly, deposition in GenBank, and publication of central paper. As of Dec 2008, the only access to the genome is through JGI Blast. The genome is small at 240 Mb and distributed across 10 chromosomes.
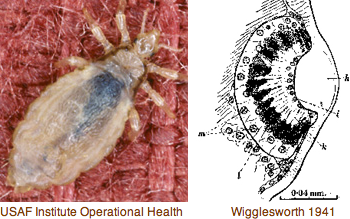
This is a subsurface deposit feeder associated with organic-rich mud, seemingly not conducive to an extensive visual system. However an extensive 1993 study of both larval and adult eyes was published in the now-defunct Journal of Morphology (online acces $25). Developing larva hava a pair of eyespots consisting of one sensory cell, one pigment cell, and one support cell. The photoreceptor cell has an array of parallel microvilli with cisternae. It is surrounded by a diaphragm formed by a pigment cell ring of microvilli-like structures. These last but a few days because at metamorphosis the larval eyespots are greatly reduced. Adults have one pair of eyes built of 2-3 pigment cells and one sensory cell in juveniles increased by 2-3 more in adults.
Unusual morphological aspects of Capitella eyes can be placed within the overall context of photoreceptor cells and eyes in Annelida, whose ultrastructural issues were carefully reviewed by Purschke in an off-PubMed journal "Arthropod Structure & Development" v35:4, 2006 (viewing issue full text costs $175). In addition to rhabdomeric and ciliary types, less-known phaosomous photosensory cells are discussed. Phaosomes (Greek: phaos = light, soma = body) were first described in the earthworm dermal photoreceptors as a central intracellular cavity (phaosome) filled with microvilli but may represent a derived form. They occur at various extraocular sites such as dermus and genitalia (in butterflies). Multiple types of photoreceptors thus provide a potential role for the diversity of opsins observed in the genome.
It's clear from Purschke's review that photoreceptors require a combination of ultrastructure, transcript expression mapping, and genomics. In other words, it's necessary to account for all the opsins found in the genome. Many photoreceptors have been overlooked entirely, notably the undirected type (no pigment cell backing); many others have stalled out in controversy for lack of gene availability.
I found a number of related opsin fragments in Capitella using various queries but surprisingly no counterpart to Platynereis ciliary opsins. One, stored as MEL1_capCap, clusters consistently with melanopsins and shares two exon breaks. It may be an ortholog of the rhabdomeric Platynereis opsin. The second MEL2_capCap is more distantly related. Reliable full length genes will require a cdna program which so far is totally lacking.
Annelida: Helobdella robusta (leech) .. 2 opsins
The JGI genome project for the leech Helobdella robusta is well along with 3,168,749 traces, a very recent assembly to blast, but no cdna. The genome is fairly small at 300 Mb but does not appear reduced in terms of gene count. Fifteen unannotated 100 kbp contigs are available at the HTG division of GenBank; these do not contain opsins but might otherwise suggest gene and retroposon densities and extent of synteny retention. The genome had not been submitted to GenBank by Dec 07.
Helobdella could be considered a promising emerging experimental system because techniques such as large-scale whole-mount in situ hybridization screening, RNA interference, and morpholino knock-down are established. It's not clear however that leech retains the degree of ancestral characters as nereid polychaetes. Until a cdna program is established, it will prove very difficult to annotate complete coding genes. The nearest species with a transcript program is the earthworm Lumbricus rubellus with 19,934 ests (but no opsins).
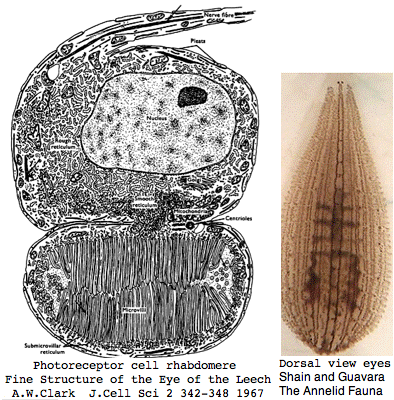
Helobdella is a rhynchobdellid, which is to say (ελεο marsh, ῥύγχος snout, βδελλα leech) a California marsh leech with a muscular straw-like proboscis in a retractable sheah for puncturing prey. Thus it is not closely related to the medicinal leech, Hirudo medicinalis. The anatomy of the closely-spaced single pair of eyes was intensively studied 40 years ago. An eye in this group consists of 30-100 photoreceptive cells in a deep pigment cup providing directional vision. Larvae are not free swimming but stay in the albuminous fluid of a cocoon. The 88 Pubmed articles include many on body plan gene expression but only two on eyes and these tangentially. We can only hope the genome project will stimulate additional studies of leech photoreceptors. It seems that every lab uses a different strain if not a different species.
I recovered two Helobedella opsin genes on 4 Dec 07 from the erratic JGI server (if no matches, close and restart with a fresh window). The full length gene, stored as MEL2_helRob has 2 conserved introns characteristic of melanopsins and its best matches there. It is likely an ortholog of a similar gene in Schistosoma, Schmidtea, Capitata, and Platynereis. The 231 aa fragment stored as MEL2_helRob has best match to octopus and chordate melanopsins and shares the first (and possibly second) intron position and phase with them. The parent scaffold 39 may contain tandem opsins or alternatively represent a misassembly. No counterpart to the ciliary opsin of Platynereis emerged. That gene -- which must have been present in the common ancestor with annelid -- could have been lost or is simply missing from the current assembly.
Mollusca: Aplysia californica (sea hare).. 2 opsins
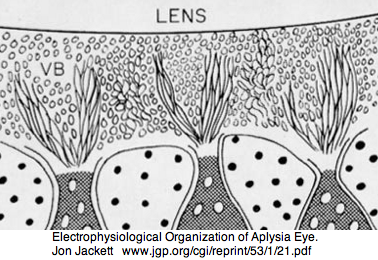
Aplysia has a pair of cephalic dorsal pit eyes just anterior to the rhinophores. The eyes are quite small at 600 microns diameter, with a spherical lens and a tiny one square millimeter retina with approximately 7000 rhabdomeric photoreceptors. Despite a fair number of studies of eyes and rhinophores involved in vision, circadian rhythm and phototactic head-waving, the opsins have not been characterized beyond immunoblot (positive for etinal photoreceptors, rhinophores, cerebral ganglia and ventral abdominal ganglia giant cell R2). There is evidence for G protein alpha subunits Gq, Gi, and Go families, phospholipase C, and an inositol 1,4,5-trisphosphate receptor in the rhinophore but this may be for chemoreception.
The sea hare genome has recently be sequenced by Broad Institute. Sizeable assembled contigs are now open to tblastn at the "wgs" division of GenBank (which allows the exon pattern to be extracted). Despite the assembly, sequencing continues: 212,159 new traces were added in the last week of Nov 07. This illustrates the need to always check the primary data repository when a gene seems missing -- millions of traces might not be used in the assembly. However a close-in query is needed to get a match.
I located the first known Aplysia opsin in the 20874 bp contig AASC01108363 on 2 Dec 2007. It had a significant expectation value (e-60) but the best match percent identity within the opsin reference collection (to fellow mollusks) was only 118/319 (36%). Otherwise the best matches are consistently vertebrate melanopsins. This gene is a strong candidate for an invertebrate melanopsin ortholog. It is stored as MOLL_MEL_aplCal.
Indeed, there are four exons but precise boundaries are difficult to locate at this low percent identity without cdna or reliably intronated guide sequence from a closely related species. However 2 introns clearly have identical position and phase to vertebrate melanosins and a 3rd quite likely; otherwise there has been intron loss in Aplysia. The contig unfortunately does not contain any information (according to blastx) on adjacent genes (synteny) despite 10 kbp still available 3'. No counterpart to the Platyerneis ciliary opsin could be found.
On 28 Dec 07 I located a full length peropsin PER_aplCal, a likely ortholog (from exon breaks and best-blast) to squid retinochome which has an excellent structural model and counterion study. The Aplysia peropsin is well-represented with 11 transcripts from pedal-pleural ganglia, CNS (adult and juvenile 1), metacerebral cells, and MCC metacerebral neurons but only terminal exons are found in the assembly. However the cdna provide a window to the trace archives which allows accurate intronation of the full gene.
It is not at all clear what relationship these lophotrochozoan peropsins have to deuterostome peropsins, nor why they seem missing altogether in ecdysozoa, nor what their ancestral status is. The 3 molluscan peropsins cluster cleanly enough with vertebrate peropsins but overlap only partially in intron placement. That could result from relatively recent intron gain and loss or reflect a much deeper ancestral splitting of peropsin classes. Representatives of these may survive more completely in echinoderms, hemichordates, and cephalochordates. Peropsin may very well be capable of ciliary opsin type signaling with trans-retinal as agonist.
At this point, Aplysia is not a Rosetta stone for opsin evolution. It is however the first mollusk with a genome assembly. This may eventually allow confident transfer of orthology validated by synteny, intron pattern, and indels. The eyes appear homologous in many aspects to those of Arthropoda supporting the common ancester of Protostomia having rhabdomeric lensing eyes, though true across-the-board homology of all eye components is a very complex subject.
Mollusca: Lottia gigantea (limpet) .. 2 opsin
The limpet Lottia gigantea was intended to be the first lophotrochozoan for whole genome sequencing but that goal slipped. It has ancestral-like spiral cleavage and trochophore larva. The genome is small relative to other molluscs at 500 mbp. Some 5.3 million traces were sequenced by May 2005. In Jan 2007 the sequencing center presented the genome at a meeting talk. However by Dec 2007 no paper had appeared. Recently JGI enabled blast of the assembly and display on their funky browser. However nothing was submitted to Genbank. JGI predicts 4 rhodopsins for its KOG gene collection; however none are recognized by the Opsin Classifier. No transcripts are available, though other molluscs have numerous ests. A German group suggests that the genome sequenced was in fact Lottia scutum.
Under these circumstances, I annotated two Lottia melanopsin in Dec 07, MEL1_lotGig and MEL2_lotGig. Their best match is to other Gq-coupled molluscan opsins, with the first probably an ortholog. Both genes have 3 exons with the two splice positions and phases identical to those of melanopsin (which in vertebrates has numerous other introns). A long run-on carboxy terminus is also seen here. It needs to be established whether these introns are ancestral generic GPCR introns or diagnostic and informative of melanopsins as a gene class. No counterpart to the ciliary opsin of ragworm was immediately apparent.
On 28 Dec 07, I recovered a peropsin, PER_lotGig, very likely orthologous to a peropsin in squid (called retinochrome there) and Aplysia (PER_todPac, PER_aplCal). Extensive structural and experimental evidence is available for squid which likely transfers over, notably the Glu181 counterion proposed ancestral. The Lottia and Aplysia peropsins are intronated identically and by inference the squid. However these differ in some respects from chordate peropsins, suggesting either intron gain or loss or alternately a small 'cloud' of ancient peropsins that were intronated slightly differently in early metazoa.
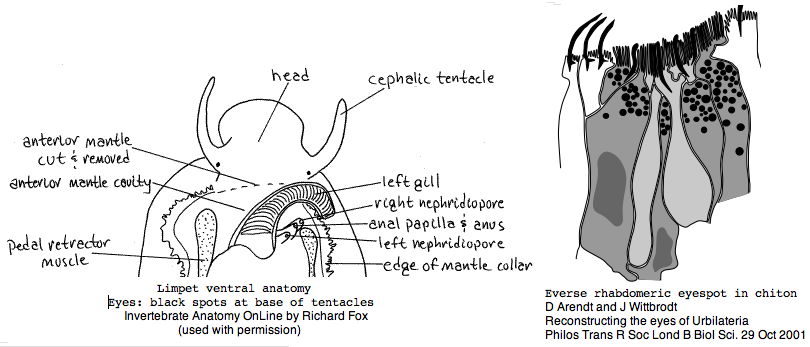
Lottia is not emerging as a model organism. There are only a handful of studies at PubMed and none on vision. The adult limpet has a pair of eyespots at the base of its cephalic tentacles that likely house a rhabdomeric opsin, perhaps the one annotated here. There may be a second role for paired eyespots in the free-swimming larva for those five days (thoroughly reviewed for chiton trochophores by Arendt and Wittbrodt but not Lottia specifically). Circadian rhythm might involve an additional opsin. The adult is an algal gardener that clears and defends intertidal areas -- raiding limpets are sensed (visually?) and driven off. The opsin sequence found here, stored as MOLL_MEL_lotGig, suggests rapid divergence rather than living fossil character. However patellogastropods such as Lottia with symmetrical non-coiled, conical shells are sometimes taken as ancestral form.
Platyhelminthes: Schmidtea mediterranea (planaria) .. 1 opsin
The common planaria Schmidtea mediterranea has a 865 Mb genome very recently assembled from 17 million traces to 10x and placed in the wgs division of GenBank, after an initial impasse attributable to high AT (69%), repeat content (46%) and high clonal heterozygosity. The genome project is described in a white paper and has a dedicated site SmedDb. It has a strong EST collection as well.
The planarian central nervous system consists of a bilobed brain and two longitudinal ventral nerve tracts connected by commissural neurons. When planarians are decapitated they can completely regenerate a new brain, including new eyes, a boon to opsin research. The structure of the eye had already been described by 1915. Regeneration of the nervous system is a very active research area.
I began with various fragmentary opsins and ESTs and recovered a nearly complete melanopsin (including all introns) from trace archives. It is stored at the Opsin Classifer as RHAB_schMed and discussed in the Schistostoma section as a likely ortholog. Since the site of expression is known from hybridization and no other Schmidtea opsins are apparent, this is likely the principal photoreceptor both here and in Schistostoma. No counterpart to the Platynereis ciliary opsins can be found in the current assembly, indicating (since they could hardly have been invented in Platynereis) their loss in Platyhelminthes is a derived condition.
Platyhelminthes: Schistosoma mansoni (trematode) .. 3 opsins
The blood fluke Schistosoma mansoni is a major agent of schistosomiasis (bilharziasis), infecting more than 200 million people worldwide, with the fresh water snail (Biomphalaria glabrata -- a large EST project) as intermediate host. As an endoparasite residing deep inside lungs, hepatoportal circulation, and mesenteric veins, it would not seem a promising species for eyespots or even circadian rhythm opsins. However at least two life stages are affected by light: the hatching of the miracidium from the egg and emergence of cercaria from the snail. These swim upwards to the surface of the water and are also affected by shadows and turbulence.
GPCR proteins are the target of approximately half of all pharmaceuticals. For that reason, a Schistoma opsin came to be studied. That gene is expressed in the miracidia and cercaria stages but down-regulated in the adult. Expression is localized to sub-tegumental structures at the front end of cercariae. Full text of the 2001 article remains locked behind a sick commercial firewall, as does a 1975 electron microscopy study of photoreceptor lamellae seen as extensions of modified cilia.
Version 4.0 of genome is readily available for blast though it is missing from GenBank as are two million of the 3.8 million total traces (7x) despite NAID funding. It's unclear whether the extensive EST set of 31000 assembled sequences is available there. The Schistosoma genome is approximately 270 MB with low GC content 34%, moderate retroposon levels andwith an estimated 15-20,000 coding genes.
I determined the intron structure of the published opsin gene (called MEL1_schMan in the opsin classifier) which classifies with melanopsins. Using this as probe, a second full length paralogous opsin MEL2_schMan was annotatable. While percent identity was only 46%, the intron structure and alignment classification were identical. Possibly this second gene has a role in the miracidium, though the first gene is expressed in both stages, more compatibly with "two color non-imaging" eyes. MEL3_schMan is similarly intronated and fairly diverged.
The first opsin is more closely related in sequence to the sole known opsin in Schmidtea, RHAB_schMed where it possibly plays a homologous role. As queries, these proteins turn up closest matches at GenBank EST in other platyhelminthes. These observations do not support the notion of horizonal gene transfer of opsins from the host snail, another Lophotrochozoan which by itself might favor sequence clustering. It would be feasible to explore synteny in both platyhelminthes.
I investigated conservation of intron position and phase using the reliably intronated match with either MEL1_gasAcu of stickleback minnow (or equally MEL1a_braFlo of amphioxus). Here the percent identity is fairly low (39%) but enough patches of good matching suffice to reliably anchor the alignment. There is perfect agreement of the first three intron positions and phases, below.
This is strong evidence for a very deep connection vertical descent of these genes from a common ancestor (eg, orthology) because these introns are highly specific to melanopsin within the opsin superfamily, ie are not generic GPCR introns as seen from the total mismatch to Ixodes, Apis, and vertebrate ciliary opsins. These same introns are predicted for opsins from transcript species such as LOPH_RHO_plaDum (Platynereis dumerilii) and MOLL_MEL_patYes (scallop). It remains to be demonstrated that all these melanopsins play a conserved consistent homologous role.
Ecdysozoa: 24 opsins
This clade includes insects and other arthropods but not molluscs and annelids (lophotrochozoa). The focus here is on species with genome projects that allow complete opsin repertoires to be determined, as supplemented by annotation transfer from experimental species when 1:1 orthology can be established.
Genome projects have not sampled ecysozoan phylogenetic diversity evenly to date but that may change as small genomes can be rapidly sequenced today. Studies of photoreception in non-genome species are limited by their inevitably incomplete repertoire of sequenced opsins and companion genes. Opsins in genomic species have determinable intron positions and phases and flanking genes so better prospects for inference of accurate descendent relationships.
An immense amount of experimental work on Drosophila melanogaster, recently reviewed from an evolutionary perspective, provides an excellent understanding of the evolutionary history underlying regulatory genetics, biochemistry, developmental and structural homologenization of opsin expression across larval Bolwig organs and adult ocelli and eye.
While annotation transfer to the other 11 fruit fly genome projects is largely justified, that becomes problematic even across Insecta because of gene loss in drosophilids (notably all ciliary opsins), lineage-specific tandem expansion of opsin multiplicities and the necessary rationales for their retention, derived conditions, and better representation of ancestral characteristics in other species. It will prove very difficult even to get at ancestral dipteran vision starting from Drosophila. Yet species with simpler vision like Tribolium are no living fossils either, having lost opsins.
Imaging vision in ecdysozoa (and lophotrochozoa) is quite different from the chordate system, with rhabodomeric opsins residing in specialized microvilli rather than ciliary opsins in modified cilia. The signalling system and chromophore regeneration also represent substantial departures. At first there seems no common ground for a shared Ur-bilateran ancestor -- which signalling system was originally used for imaging vision and which lineage displaced it with the other? Some protostomes still utilize ciliary opsins in non-imaging photoreception and similarly some deuterostomes still utilize rhabodomeric opsins. Since the relevent opsin gene trees coalesce far earlier, this proves Ur-bilatera possessed both opsin classes (without clarifying which system was used for imaging vision, if either).
Blastp of any rhabdomeric opsin from any protostome against the set of all deuterostome opsins invariably gives vertebrate melanopsins as best match, whereas blastp of any protostome ciliary opsin (pteropsin) always has best match to TMTs (ancestral form of encephalopsin). That is, from the biomedical perspective, rhabdomeric opsins are just a clade-specific expansion of melanopsins largely irrelevent to human vision. Similarly invertebrate ciliary opsins not used in imaging vision primarily inform us on deeper ancestral origin issues. Note melanopsin and TMT are not orthologs at the level of Ur-bilateran nor even Ur-eumetazoan because gene duplication and divergence preceded the cnidarian last common ancestor.
The nature of vision at ancestral nodes has not yet been resolved, in part because pre-bilateran cnidaria photoreceptors studied so far as outgroup have been either ciliary, or based on distantly related cnidarian-specific opsins, or in the case of coral melanopsin, genomic sequence not yet associated with photoreception. In the Ur-eumetazoan common ancestor, this could imply ciliary opsin imaging vision, no imaging vision but convergent evolution (later independent invention) in the box jellyfish lineage, or even rhabdomeric imaging vision with subsequent displacement by ciliary opsins in cubomedusa and separately in later deuterostomes. Sponge larva presumably also utilize a ciliary opsin but here again it is unclear whether later metazoan use a system descendant from that.
It's sometimes asserted that imaging vision systems (all highly dissipative of ATP) were first enabled in the rapidly oxygenating Cambrian ocean, yet near-simultaneity is not a good fit to the arthropod fossil record (stalked eyes) nor molecular reconstructions. For example, extant representatives of early diverging deuterostomes (xenoturbella, acornworms, echinoderms, tunicates, amphioxus) all lack imaging vision (depending on how that is defined in scanning larva), so it seems clear that early arthopods had well-developed vision prior to the emergence of hagfish/lamprey. The majority of extant animal phyla have prospered for 540 myr without ever developing imaging vision.
Ecdysozoa .. opsin repertoire of the last common ancestor
Questions of ancestral opsin repertoires and their implied photoreception biology are best addressed after careful step-by-step reconstruction of opsin repertoires in each of the relevent lineages, exhausting available information in extant species rather than just add to a century of speculation. These reconstructions can help evaluate candidates for 'living fossils'. Thus the focus in this section is reconstruction of the opsin repertoire of just the last common ancestor of ecdysozoa. That can be combined later with parallel efforts on ancestral lophotrochozoa and deuterostomes opsins to get closer to the Urbilateran.
This program has already been set in motion with important recent papers sequencing opsins in arthropod outgroups to the over-sampled Insecta, providing new opsins from crustacean and chelicerates. The gene tree, as overlaid on clade divergence, shows color vision already well established prior to divergence of insects. However incoming data, in the form of a fifth opsin from the ventral eye of horseshoe crab Limulus (a chelicerate, not crab (malacostracan crustacean)), already requires an earlier origin for the opsin class BcRh1 once thought specific to crustaceans.
Just as absence of effort on hagfish has needlessly delayed our understanding of chordate vision evolution, absence of effort on early diverging Ecdysozoa such as Pycnogonida (sea spiders), Onychophora (velvet worms), and Tardigrades (water bears) has seriously retarded reconstruction of the ancestral opsin repertoire in this lineage. Rather than yet another obscure mammalian cone opsin or yet another butterfly gene expansion, biology is better served by more strategically placed species. Ironically the truly pivotal data may come out of genome projects rather than opsin research per se.
In classifying ecdysozoan opsins from a deeper evolutionary perspective, it is necessary to set aside narrow clade-specific expansions and contractions of opsin repertoires, however adaptively important to the individual species concerned. Wavelengths of peak adsorption -- subject to significant change from tuning residue substitution -- seem an unsound basis for evolutionary classification (though in retrospect work fairly well). This leaves phylogenetic alignment, signature residues and rare genomic events (such as indels and introns) as the main tools.
Here the remarkable observation in 2003 that a single lysine K90 (bovine rhodopsin numbering G90) suffices to define the phylogenetically valid class of ultraviolet opsins. Six years later, despite a vastly expanded data set, there is still perfect concordance of spectrophotometry, behavioral studies, alignment, signatures, gene structure, and possession of lysine at this position. This residue was previously known to be important to spectral tuning from bird C90S ultraviolet vision and human rhodopsin G90D night blindness.
This residue sits deep within transmembrane helix 2. That hydrophobic mileau is unworkable energetically for positively charged lysine unless a compensatory counterion exists. That negatively charged residue is presumably the ancestral counterion, negatively charged E171 (rather than the E113 of vertebrate ciliary opsins). K90, by taking E171 away from the Schiff base lysine K296, has the effect of leaving that protonated, an effect known to shift adsorption into the ultraviolet.
Observe however that opsins specialized to blue (not ultraviolet) are also satisfactorily classified in this same region (June 2009 current alignment below). These opsins have some other residue than lysine at 90 but share a one-residue deletion near the lysine that would shift its orientation relative to the chromophore as well as a proline six residues after the DRY motif, which is glycine in all other ecdyozoan opsins. This agrees with conventional phylogenetic alignment that sisters blue and ultraviolet opsins to the exclusion of long wavelength and blue-green opsins as well as to the more basal BcRh opsins operationally defined by clustering to two particular opsins from the crab Hemigrapsus sanguineus.
While K90 might have arisen multiple times as the same solution to the problem of ultraviolet vision, the simultaneous presence of multiple other defining signatures render this improbable. Opsins with K90 thus date back to the common ancestor of chelicerates and insects (ie Arthopoda) if not earlier, though no such opsins are seen in lophotrochozoan whole genome projects (eg the mollusk Aplysia) or deuterostomes. Blue optimized opsins appear limited to insects. Consequently prior to gene duplication and divergence, the ancestral gene had K90, hence ultraviolet vision not tuned to blue.
Panarthropoda: Hypsibius dujardini (water bear) tardigrade 0 opsins
A 5x genome project for Hypsibius dujardini, a phylum of microscopic ecdysozoan was approved in July 2007 but Broad has not yet begun trace reads on the small 70 mbp genome (suggesting densely spaced genes with small introns as this is not likely highly derived). It could prove very useful for opsins as tardigrades are basal to all of Arthropoda and so shed light on that last common ancester. In fact with accompanying centipede, horseshoe crab, amphipod, and priapulid genomes, the whole ecdysozoan ancester will be accessible.
The only known fossil specimens are found in Siberian mid-Cambrian deposits and much later amber. The older fossils have three pairs of legs rather than four, a simplified head morphology, and no posterior head appendages and probably represent a stem group of extant tardigrades. Aysheaia from the Burgess Shale might be related to tardigrades.
Nothing is currently known about photoreception or opsins in tardigrades -- or even if they have eyes. However it looks like we can expect some rhabdomeric opsins at the minumum in front of these pigment cups. However the current GSS and EST collections (about 6000 sequences) do not currently contain any convincing matches using various rhabdomeric and ciliary opsins as tblastn queries. Tardigrade photos and movies provided by Goldstein Lab
Chelicerata: Ixodes scapularis (tick) 2 opsins
The genome project was completed long ago but has experienced a multi-year bottleneck in assembly release and publication. However contigs built from a subset of 19.4 million traces became available to tblastn of the GenBank "wgs" division by late 2007. Ixodes has a very conservative genome (regretably 2.1 gbp in size), seemingly far less derived than drosophilids in matters such as intron, gene retention, and protein sequence conservation. This, in conjunction with the helpful phylogenetic position of chelicerate outgroup to the many insect genomes, has improved prospects for reconstructing the ancestal opsin repertoire of Arthropoda and eventually Protostomia and UrBilatera.
A large collection of annotated Ixodes ESTs is available at the DFCI Gene Index of which 3 are marked up (2 wrongly) as opsins. Using the Opsin Classifier, the full length gene could be recovered for the first of these (TC19272) on 24 Nov 07, intronated at the Trace Archives (4 introns, superb coverage), and added to the classifier fasta collection as RHAB1_ixoSca. It classifies with rhabdomeric opsins (ie with deuterostome melanopsins) with a very respectable 57% maximal percent protein identity. The second and third intron have classical ancestral position (following GWSR and LAK) and phase (2 and 0). Synteny awaits assembly of large contigs -- adjacent exons are not spanned by single traces. An apparent ciliary opsin fragment in Ixodes was located using that of Platynereis dumerilii as probe, it is stored as CILI_ixoSca but needs further analysis.
A fragment from a second opsin, found in June 2009, consists of a single long exon, enough to contain the key opsin motifs ERY...KSAAVLDPFVYGLSHPSFR. This opsin classifies most closely (44% identity) with UV opsins from Daphnia, Pediculus, and Apis. This gene fragment could not be extended because assembly contigs are so short . It is not retroposed because the upstream splice acceptor AG is retained in expected position and phase. Ixodes thus appears to have a small repertoire of opsins.
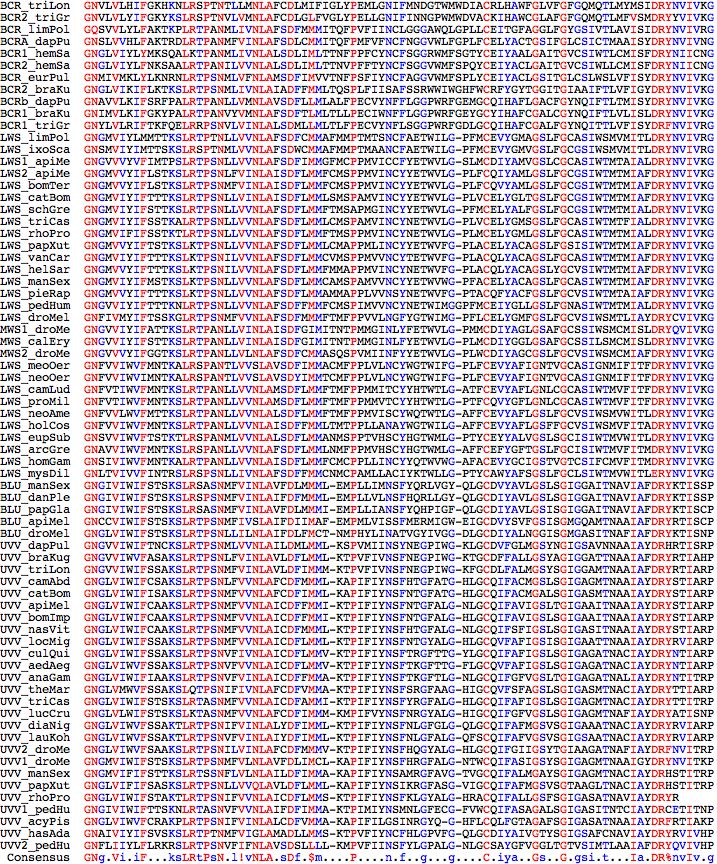
The 11 chelicaterate opsins available in June 2009 (after consolidating nuisance GenBank entries for Limulus): BCR_limPol Limulus polyphemus (horseshoe_crab) opsin 5 FJ791252 ventral eye LWS_limPol Limulus polyphemus (horseshoe_crab) CRBOPSINA L03781 lateral eye LWS1_hasAda Hasarius adansoni (jumping_spider) HaRh1 kumopsin1 AB251846 LWS2_hasAda Hasarius adansoni (jumping_spider) HaRh2 kumopsin2 AB251847 LWS1_plePay Plexippus paykulli (jumping_spider) PpRh1 kumopsin1 AB251849 LWS2_plePay Plexippus paykulli (jumping_spider) PpRh2 kumopsin2 AB251850 LWS_ixoSca Ixodes scapularis (tick) P35361 ocellus LWS_loxLae Loxosceles laeta (spider) fragment EY188471 venom gland UVV_hasAda Hasarius adansoni (jumping_spider) HaRh3 kumopsin3 AB251848 UVV_plePay Plexippus paykulli (jumping_spider) PpRh3 kumopsin3 AB251851 UVV_ixoSca Ixodes scapularis (tick)
The phylogenetic arrangement of these species is (Limulus,(Ixodes,(Loxosceles,(Hasarius,Plexippus)))). Molecular clock dating of divergences (late Paleozoic just for land chelicerates) is under some dispute.
This leaves Pycnogonida (sea spiders) the last major unrepresented chelicaterate group (even more basal if chelifore appendages aren't homologous to true chelicerae, in conflict with Hox expression boundaries showing anterior-most appendages also deutocerebral). Shallow water species have two pairs of dorsally located eyes. Given that the body is generally just a millimeter or two, these eyes are small and quite simple. A longwinded 1891 dissertation on their larval and adult anatomy is available as well as a 1973 ultrastructural and modern account.
The arthropod outgroup Onychophora are also completely lacking in opsin data even though their eyes may provide important clues to the evolution of arthropod rhabdomeric vision -- a pair of simple ocelli may be the ancestral visual design as in the basal Onychophora.
Crustacea: Daphnia pulex (water flea) .. 7+ opsins
An 8.7x genome assembly was released in July 2007 at JGI with further support at wFleaBase. A May 2009 meeting report suggests an imminent release of initial publications by the 370-member consortium.
The gene count, supposedly 39,000, may be inflated with genomic transcript noise that does not really code for protein, contig assembly errors resulting from polymorphism and use of paired end reads and over-counting of gene fragments and recent processed pseudogenes. JGI and Gnomon models to date err grievously on ciliary opsin gene models because they lack the last exon (below) which is necessary to complete the covalent lysine motif to FR.
This crustacean, basal to Hexapoda arthropods, provides a potentially important outgroup to insects (together forming Pancrustacea). However the opsin story, summarized in a meeting abstract is an embarrassment of riches, not conducive to deducing ancestral arthropod genome content. The total number of opsin genes came in at 37, comprised of 22 rhabdomeric opsins (mostly long wavelength), 7 ciliary opsins (pteropsins), and 8 in a novel family without close affiliates. A post on the Ixodes list serve even raises this to 46 by Feb 2008.
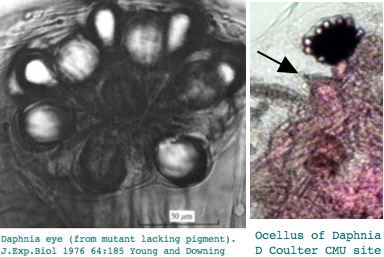
This seems excessive given Daphnia has a single medial compound eye with merely 22 ommatidia with 8 photoreceptors each, an under-focusing lens, and a three-ocellus naupliar eye, yet circadian rhythms and a need to assess water turbidity, depth, and distance fkom shore. Daphnia also can detect polarized light. It's not clear that exquisite color discrimination potentially afforded by dozens of opsins would be advanageous for a 22-pixel array; experimentally, only four wavelengths of peak sensitivity are observed at 348 (UV), 434, 525, and 608 nm in dorsal ommatidia.
Again the possibility arises that K-rhodopsin gene duplicates could have taken on other sensory or metabolic roles (digestion of complex algal carotenoids). Planned in situ hybridization studies may illuminate biological roles of these opsins. The pteropsins are probably of most interest from the urbilateran perspective.
Gene models have not been submitted yet to GenBank but are extractable by text query at wFleaBase. What is needed here however is not the clutter of 37 sequences but their collapse into UV, blue, long, pteropsin, and novel ancestral representatives. This would remove the noise from lineage-specific expansions. The intron structure could provide very important support to classification schemes.
To a certain extent, this has been accomplished by June 2009 at FleaBase as text searching by 'opsin' turns up 25 matches, many in tandem pair sets (which could reflect assembly error to some extent). There is no explanation of how 37 opsins got expanded to 46 then reduced to 25 with ciliary and novel opsins no longer not listed. Despite assigned accessions, no gnomon gene models have been released at NCBI.
Intronated gene models can be manually extracted from scaffold dna (done for four below). These models, taken at face value, unsurprisingly have best-blast at GenBank to Triops and other crustaceans (20 non-Daphnia opsins, all melanopsins), which mercifully have been analyzed in a careful Feb 2009 paper. This study considered only non-EST Branchiopoda (like Daphnia) and Malacostraca melanopsin sequences that likely under-represent opsin evolutionary information available from the full seven classes of Crustacea.
The only Daphnia opsin (NCBI_GNO_472553) with a transcript (FE295533) has been assigned to the BCRH1 group (middle wavelength MWS). One Daphnia opsin has a lysine at position K90 (bovine rod rhodopsin numbering) considered proof of UV purposing.
The value of Daphnia genomic opsins relative to other crustaceans lies in their intronation, which distinguishes expansions arising through retroprocessing from tandem and segmental duplication of a few master intronated genes (which would then be the orthologs to other arthropod opsins).
Indeed the intronation pattern -- typically far more deeply conserved than protein sequence -- could link pteropsins more convincingly to lophotrochoan and deuterostome opsins than alignments with percent identities in the 20's. However, in comparison to Apis opsin counterparts, Daphnia has experienced numerous intron gains and losses, not furnishing a good guide to the ancestral state.
NCBI_GNO_176434 scaffold_53:626704-628972 Blue opsin [probable ortholog of Triops longicaudatus RhC] NCBI_GNO_416624 scaffold_95:369266-373273 Opsin Rh3 Inner R7 photoreceptor cells opsin NCBI_GNO_366144 scaffold_14:844292-847788 Melanopsin NCBI_GNO_557324 scaffold_2568:2224-6662 Short wavelength-sensitive opsin [defective model fragment but KMAACVDPFVYAINHPKYR] NCBI_GNO_750363 scaffold_40:707906-709794 Compound eye opsin BCRH1 (brachyuran crab RH1) NCBI_GNO_754363 scaffold_40:716143-718346 Compound eye opsin BCRH2 (brachyuran crab RH2) ... (rest are BCRH1 and BCRH2 types) >UVV1_dapPul Daphnia pulex NCBI_GNO_176434 FE384049 EST 53% identical Apis mellifera 69% Triops RhC 0 MLGWNTPEDYMSYVHP 21 YWKTFEAPNPFLLYMIGFLYTIFMFCCVAGNGVVIWIFTN 2 1 CKSLRTPSNMLVVNLAILDMLMMLKSPVMIINSYNEGPIWGKLGCDVFGLMGSYNGIGSAVNNAAIAYDRHR 2 1 TISRPLDGKLSRKQVTLMIVAIWAWATPFSVMPFLGIWGRYVP 1 2 EGFLTTCTFDYMTEDASTRFFVGSIFVYSYVIPLAMLIFYYSKIVRSVGDHEKTLRDQAKKMNVTSLRSNRDQNEKSAEVRIAKVAIALATLFVFAWTPYAFVALTAAFGNR 2 1 SVLTPLLSTVPACCCKLVSCINPWIYAINHPRYR 2 1 MELQKKMPWFCIHEPVPTNDDSSVGSATTEMSGVSKETSS* 0 >UVV2_dapPul 49% penultimate intron lost, last intron has slid back 2 aa 0 MNGWNTPADYKSYVHPHWLSYEEPNPMLHHLLGVLYIFFMIASCLGNGIVIYIFST 2 1 TKELKTPSNILILNLAICDFIMMIKTPIFIVNSFNEGPVFGRLGCSIFGLLGAYVGPCSAVTNAAIAYDRYR 2 1 CISDPMGKRWSKSQASLIVLGCWVYASPVSLLPFTEIVNRFVP 1 2 EGYLTSCTFDYMTDNLETKMFVFILWIWCWIMPLGVIIFSYGKITTQVMTHEARLKEQAKKMNVESLRSGANKDARNEIRVAKVGISLTTLFLLSWTPYFAIAFIGCYGNR SLLTPGLSMIPACTCKMAACVDPFVYAINHPK 2 1 YRLELMKRFPWLCVHEKDDSTRSENSTNATIASEAESRT* 0 >BCRa_dapPul Daphnia pulex NCBI_GNO_149114 53% identical MWS_hemSan, 72% Triops longicaudatus RhA AB293433 0 MSNNLSSGYSSVAYRSEGASVLWGYPPGLSIVDLVPDDMKEFIHPHWNKFPPVNPMWHYL 21 LGVIYVILGITSVT 1 2 GNSLVVHLFAKTRDLRTPANMFVINLAFSDLCMMITQFPMFVFNCFNGGVWLFGPLFCELYACTGSIFGLCSICTMAAISYDRYNVIVNGMNRRRMTY 1 2 GRAGGLILFCWIYAIGWSIPPFVGWGKYIPEGILDSCSFDYLTRDTM 0 0 TISFTCCLFAFDYCVPLIIIIFCYYHIVRAIVHHEDALRDQAKKMNVSSLRSNADQKSQSAEIRVAKIAMMNITLWVAAWTPYAAICLQGAVGNQDKITPLVTILPALIAKSASIFNPVVYAISHPKYRL 0 0 ALQKALPWFCIHEKEEKEPPQDRREDSQSIATTNTNSSDVSLP* 0 >MEL1_dapPul Daphnia pulex NCBI_GNO_366144 no close homologs 0 MTSSNDSAGYLWAINATIWIIDDSNETLGIDWDDWDVSLWTQEQRQLLEHGGIPRQVHVALGVLLSFIVLFGFAANSTILYVFSR 2 1 FKRLRTPANVFIINLTICDFLACCLHPLAVYSAFRGRWSFGQT 1 2 GCNWYGMGVAFFGLNSIVTLSAIACERYIVITSSSCRPVVAKWRITRRQAQK 0V 0 VCAGIWLHCAALVSPPLLFGWSSYLPEGVLVTCSWDYTSRTLSNRLYYFYLLFFGFFLPVSVLTFCYAAIFRFILRSSKEITRLIMTSDGTTSFSKSTVSFRKRRRQTDVRTALI ILSLAILCFTAWTPYTIVSLIGQFGPVDEDGELKLSPMVTSIPAFLAKTAIVFDPLVYGFSSPQFRNSVRQILRQQSISSSGNAGNRAGPNNMAMARTAIQNSRASSHATVSSF SRNARMFPKDPLSKKTPNDPFVSTPLAVQQIPHFRLPTDVDINEQQFRRGIYANKSVSYWIDIIVLLQLGENLRKSCMKRKNSFKIPAGSIPQKNKLSNSRCSLLEDVSTHSLA LRQMIFRKEGELYLFHHQPSHNAELAANKMDHQGNNKRIRRRFSEADMMHRSGKCRKNLPVSTSFDQ* 0 Daphnia opsins have no experimental data but their 'best-blast to PubMed' allows inference from opsins with experimental data: UVV1_dapPul NCBI_GNO_416624 ... 43% Acyrthosiphon pisum rhodopsin 7 XM_001944891 BCRH1_dapPul NCBI_GNO_149114 ... 72% Triops longicaudatus AB293433 MEL1_dapPul NCBI_GNO_366144 ... 33% Patinopecten yessoensis scop1 Gq AB006454 TMTa_dapPul ... 36% Apis mellifera pteropsin TMTb_dapPul ... 36% Apis mellifera pteropsin
This blast twilight zone is especially dangerous for photoreceptor opsins because they are embedded in much larger gene family of generic rhodopsin and GPCR which share many structural and signaling properties. A slowly evolving generic rhodopsin might well score higher than fast evolving photoreceptor opsins. Gene expansions are noted for markedly enhanced rates as copies neo- or subfunctionalize. The generic rhodopsin might also share diagnostic residues through convergence at least at the level of statistical signficance ambiguity. Consequently intron location/phase and synteny can provide important backup.
The synteny circle surviving at this phylogenetic depth will be local (optimistically Pancrustacean). That is, the blue opsin of Daphnia might in synteny with Drosophila (ie establish orthology) but not to Platynereis ciliary opsin much less any vertebrate opsin (eg encephalopsin). This could be remedied to some extent by ancestral gene order reconstruction. The degree to which synteny can contribute to validating orthology relations within opsins is not currently known.
Ciliary opsins for Daphnia, absent from the collection of 25 pipeline-labelled genes, can be located by querying with Anopheles counterpart. Stored at the Opsin Classifier as TMT_dapPul, these are plausibly orthologs of deuterostome and lophotrochozoan ciliary opsins, as are new ciliary opsins from Culex, Aedes, Tribolium, and Bombyx. Counterparts to this gene and presumably its associated photoreceptor structure are missing in Drosophila, Nasonia, and other genomes.
In Daphnia, with its high level of apparent tandem duplication and 'excess' of opsins, the opsin of each class with highest external blastp score may be the parental gene and best conserve the function observed in its counterpart in other species.
>TMTa_dapPul Daphnia pulex (water_flea) last exon uncertain 45% id TMT1_anoGam 0 MPVWVYWSASAYLLFISIAGLFMNIVVVVIILNDSQ 0 0 KMTPLNWMLLNLACSDGAIAGFG 2 1 TPISAAAALKFTWPFSHELCVAYAMIMSTA 1 2 GIGSITTLTVLALWRCQHVVWCPTNRNSNFTDPNGRLDRRQGALLLTFIWTYTLIVTCPPLFGWGRYDREAAHIS 2 1 CSVNWESKMDNNRSYILYMFAMGLFIPLMAIFVSYISILLFIHK 0 0 SQQTSNNSDTVEKRVTFMVAVMIGAFLTAWTPYSIMALVETFTGDNVTNDSVSSEIKFYAGTISPAVATVPSLFAKTSAVLNPLIYGLLNTQ 0 0 FRTAWEKFSSRFLGRKKRHQRSQMAMGVSHKRRRDYLRTLLNRPASDEPAIVQHPSTKEMASSQAVSCVVVSNLDVPRAPNNSYVTVNDE* 0 >TMTb_dapPul Daphnia pulex (water_flea) ciliary long tail 60% identity 0 MPTWAYRLTAAYLLLISVLGLIMNVVVVIVILNDSQ 0 0 RMTPLNWMLLNLACSDGAIAGFG 2 1 TPISTAAALEFGWPFSQELCVAYAMIMSTA 0 0 GIGSITTLTALAIWRCQLVVCCPAKRKSAFTNHSGRLGCRQGVILLVIIWIYALAITCPPLFGWGRYDREAAHIs 2 1 CSVNWESKTNNNRSYILYMFCMGLVVPLAVIIISYVRILRVVQK 0 0 NQQQSGNVHRHRRDAAEKRVTMMVACMIAAFMAAWTPYSILALFETFIGQDNHSTYYSSRINNATNFSSAFPDGDLSYVGTISPAFATIPSLFAKTSAVLNPLIYGLLNTQ 0 0 FRLAWERFSLRFLGRFQCHRTQGVSGQHGANHHKTRRNVRKYLPNCYGDSRSLKPTPTVHLPMKEMVVSHAEQKVKTAQEQASSSVTKITTIPLISSDNQTIVSCPSSIMAN CQQHETNQANHQQAARPDKVVDHQHLLQPNRLSSLLSLSLPSVLISTPNLPCSAQRQSAAEDQAMATCQQMTSGRIRDQQQQSDSFVVVGLLSRSADCYHHHTGDVEQFVFLDSTVDELGLTARSASP* 0
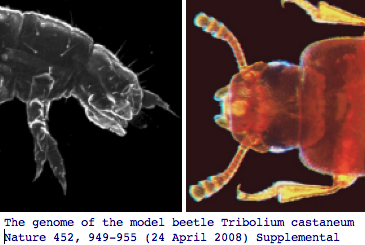
Panarthropoda: Tribolium castaneum (flour beetle) .. 3 opsins
The red flour beetle, which is highly dark-adapted in lifestyle, has lost its blue opsin according to both the newly published genome project and specialized experimental querying, retaining the other two ancestral color vision opsins and encephalopsin (which is called pteropsin in insects though likely a strict ortholog). The Tribolium genome article 110 page supplemental contains an excellent Table S14 of all known genes involved in insect eye development.
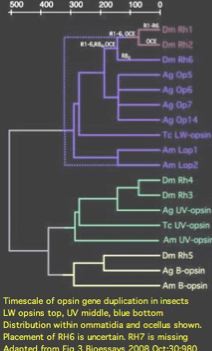
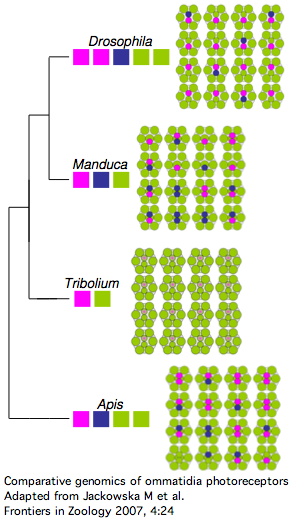
Insect opsins are expressed non-uniformly across individual eye units (ommatidia) within compound eyes. In Drosophila, six peripheral photoreceptor cells R1-R6 express LW opsin which detect brightness, projecting into the upper optic neuropil (lamina). Central photoreceptors R7 and R8 provide color vision via UV, blue, and LW opsins that project into the second (medulla). The dorsal rim area ommatidia are modified to detect polarized light.
The comparative genomics of ommatidia number and opsin utilization is indicated in the figure. Opsin gene loss raises different issues, namely replacement, from the more familiar gene gain issues (differential rewiring). After discussing various sequential mutational scenarios and the necessity of each step being adaptive or at least near-neutral, Jackowska et al settle upon expansion of LW opsin expression into all photoreceptor cells, resulting in co- expression with blue opsin in some R8 cells and UV-opsin in R7cells. This is followed by loss of expression or pseudogenization of blue opsin. Although co-expression defeats the purpose (via spectral summation) of separate opsins that enable color vision, there are precedents in butterflies and (typically nocturnal) vertebrates.
It's also known how Apis and Manduca (also genome project species) end up with nine photoreceptor cells per ommatium instead of eight -- it's due to duplication of R7 cell fate (across all ommatidia). That raises the interesting question of whether such cell duplication simply results in duplication of opsin expression at the molecular level. That's not the quite the case today because the two central R7-like cells exhibit differential opsin expression. It's not known whether additional mutations were needed to attain this.
In summary, insect genomes are fairly straightforward in terms of their contribution to establishing the ancestral arthropod visual system, but their real value lies in the extensive comparative data available within Insecta, ecological studies of adaptive vision, and the experimental genetic opportunities within Drosphila (eg a recent article exploring deviations from ommatidia expressing but a single opsin). However no single insect genome can serve all purposes because of gene loss (eg ciliary opsins in Drosophila).
That's also the case for non-opsin GPCR which have gained a new importance given the possibly paraphyly of the opsin gene tree (ie some opsin gene duplicates may have given up retinal to signal via other agonists). Here we are fortunate to have a genome-wide inventory of neurohormone GPCRs in Tribolium. This turns up 20 biogenic amine GPCR (21 in Drosophila, 19 in bee), 48 neuropeptide GPCR (45 in Drosophila,35 in honey bee), and 4 protein hormone GPCRs (4 in Drosophila, 2 in bee) with likely ligands for 45 of the 72 Tribolium GPCR. The flour beetle retains an ancestral vasopressin GPCR and cognate peptide unlike other studied insects which are not adapted to such an extremely dry environment. On the other hand, Tribolium lacks allatostatin-A, kinin, and corazonin. This covers comparative genomics of 340 million years of insect GPCR evolution -- it is very common for new agonist/receptor couples to arise and old ones to disappear. Again we see genome density sampling will need to be high to sort out Urbilatera.
Panarthropoda: Pediculus humanus (louse) .. 3 opsins
The body louse genome, being favorably small at 108 Mbp, is well along with 2.2 million traces and a contig assembly hopefully disentangled from its endosymbiont bacterium. Sequencing is medically motivated. The lifestyle of this hemimetabolous (nymph-like adult, no pupal stage) insect does not suggest a full spectrum of metazoan photoreceptors; indeed we shall find but 3 opsins. Even that seems a lot for a single lateral ocellus of 130 rhabdomeric photoreceptor cells lacking Semper and dedicated pigment cells. The broader interest here is intronation and synteny of these opsins (hence orthology), not available in many insects with opsin studies. It requires quite dense sampling to get ancestral introns for each arthropod opsin class because high rates of intron gain and loss can occur.
I reconstructed 3 multi-exon louse opsin genes on 24 Dec 07 by tblastn of numerous queries against GenBank wgs database division. These apparent rhabdomeric imaging opsins are stored in the Opsin Classifier as INSE_LWS_pedHum, INSE_UVV1_pedHum, and INSE_UVV2_pedHum. Louse otherwise seems a gene loss story in terms of relic ciliary opsins or even melanopsins so not especially favorable for retention of ancestral characters. The new opsins potentially provide trichromatic color vision to the louse in the short, blue, and long wavelength photoreception regimes, though lambda max awaits experimentation as the second ultraviolet opsin could be either re-tuned or co-opted for some other function, as in bumblebee where a UV opsin is expressed in proximal lamina rim, antennal lobe, central complex and protocerebrum clusters. That seems likely because INSE_UVV2_pedHum is back to ancestral tyrosine in (bovine rhodopsin) position E113 whereas true ultraviolet insect opsins all specify phenylalanine here (which relaxes lambda max into the ultraviolet, ie closer to that of free retinal).
CA Hill of the louse genome annotation team discussed 3 opsins back in a June 2007 email session, calling PHUM001073 perhaps an ultraviolet opsin while rejecting a fourth PHUM000074. These gene models are not released to GenBank nor is that terminology used in the meagre search capabilities of P. humanus VectorBase. Upon whole proteome file download, PHUM001073-RA turns out to be an unintronated dna fragment matching residue 44 to stop codon of INSE_UVV1_pedHum. PHUM000074-RA has nothing to do with opsins. PHUM005795-RA is missing the first 49 residues of INSE_LWS_pedHum but otherwise identical. PHUM001044-RA is a fragment beginning at residue 55 of INSE_UVV2_pedHum. In short, it's hard to find full length genes without benefit of the Opsin Classifier, cdna, or ab initio gene predictor.
Panarthropoda: Rhodnius prolixus (kissing bug) .. 2 opsins
Yet another genome project completed long ago at the trace level but sitting around unassembled until 17 June 2009. (tblastn now at GenBank). In August 2008 some 6,879,098 trace reads and 16,284 EST sequences were available. This number of traces is more than adequate for a good assembly but until now, opsins had to be fished out by exon by exon using blastn at the trace archives.
Rhodnius prolixus, a large blood-sucking hemipteran insect that is carrier for a parasitic protozoan (Trypanosoma cruzi) responsible for Chagas disease through bites around the eyes and mouth. Chagas disease is a currently incurable tropical disease that damages the heart and nervous system. Rhodnius is nocturnal, with possible implications for its opsin repertoire, but becomes active at night. It is found in South and Central America, primarily in domesticated rural areas, currently affecting 16-18 million people and killing around 20,000 people annually. Darwin is sometimes claimed to have suffered from Chagas disease as a result of a bite (implausibly in northern Argentina) reported in Voyage of the Beagle diaries.
Rhodnius clearly has three distinct melanopsins opsins and a ciliary pteropsin. The first is a long wavelength sensitive gene most closely related (84% identity) to Tribolium but whose intronation pattern is closest to Apis (a phase 00 intron is missing in Rhodnius). The second Rhodnius opsin classifies with insect UV opsins, most closely to that of Apis. The complete ciliary opsin is closest to that of mosquito and flour beetle ciliary opsins but quite diverged at 56% identity.
>INSE_LWS_rhoPro Rhodnius prolixus (kissing_bug) frag missing first and last exons 0 2 1 YQFPPLNPLWHGILGFVIGVLGIISIVGNGMVIFIFSSTKTLRTPSNLLVVNLAFSDFLMMFTMSPPMVINCYNETWVL 1 2 GPLMCELYGMLGSLFGCASIWTMTMIALDRYNVIVK 0 0 GISAKPMTNKTAMLRILLVWAFSIMWTVFPFFGWNR 2 1 YVPEGNMTACGTDYLTKNWVSRSYILVYSVFVYFLPLFTIIYSYFFILQ 0 0 AVSAHEKQMREQAKKMNVASLRSAEAANTSAEAKLAK 0^0 VALMTISLWFMAWTPYLVINYSGIFETISISPLFTIWGSLFAKANAVYNPIVYAIR 2 1 *0 >INSE_UVV_rhoPro Rhodnius prolixus (kissing_bug) fragment, novel introns, 67% identity Apis 0 0 1 ASTSGNIRTLGWNLSPEDLKHIPEHWLSYPEPEPILNYALGVLYIFFMLIALIGNGLVIWIFST 2 1 AKTLRTPSNIFVVNLAICDFLMMSKTPIFIYNSFKLGYALGHRACQIFALLGSFSGIGASATNAVIAYDRYR 2 1 ERFSTKCTFDYLTPTSEIRNFV MSLIIYFYSQIVSHVIIHEHNLREQ 0 0 AKKMNVESLRSNANMHTQSAEIRIAKAAITICFLFVASWTPYAVLALIGAYGNQ 2 1 DLLTPAVTMIPACACKAVACVDPYVYAISHPRYR 2 1 QELSKKFPWLDIKEAPAPSSVDANSTATEMTLPTQTSPAEA* 0 >INSE_UVV2_rhoPro Rhodnius prolixus (kissing_bug) Insecta; Pterygota frag 50% UVV2_pedHum 1 FRTLRTSSNILILNLAVSDFLMVAKMPVFIYNSFYFGPVLGEM 1 2 GCHFYGFIGGLSGTASILTLAAIAMDRYLGIAHPLNFNQGRAKKRTIVWITFIWVYSITFASIPLSHIGVKTYVPEGFLTSCSFDYLSTDIQNRCFIFIYFVAA WCLPLLVIITSYVGICREVLRVSLIRKGQEREQRKREAKLSAILALATFLWFLSWTPYAAVALLGIFGYKNHITQLASMIPALFCKTAACVNPFIYGLNHPRLRQQLLKLCCKKRYNLEK THFSRSWRNTSCSFKLKEQSLCNVSQSRLRRTSTVASEPSEHSTHFM* 0 >TMT_rhoPro Rhodnius prolixus (kissing_bug) Insecta; Pterygota ciliary opsin full ACPB01038514 + ACPB01038515 56% TMT_triCas 0 MLMPSAGFLAASIILFLIGFLGFFGNLIVIIIMCRDKN 0 0 LWTPVNFILFNVIVSDFSVAALGNPFTLASAIAKRWFFGQSMCVAYGFFMALL 1 2 GITSINSLTVLALERYLIVSQPVSHGSLSRPTASDIVGSIWLYSFVITiPPLVGWGEYGLEAANIS 2 1 CSINWETRSHSSTSYILFLFTFGFFIPIIVISYSYMNIILTMKK 0 0 STMNAGRVNKAESRVTWMIFVMIFAFFLAWTPYAILALMIAFFDSNVSPAIATIPAIFAKTSICYNPFIYAGLNTQVVYFFV* 0
Panarthropoda: Drosophila melanogaster (fruitfly) .. 7 opsins
Every aspect of photoreception in Drosophila has been studied for decades. Because this research is regularly reviewed at length, the focus here is on genome project developments and issues that remain in characterizing opsin function and evolution.
Drosophila has seven opsins, all of melanopsin class. Ciliary-class opsins (present elsewhere in arthropods) have been lost in all 12 drosophilid genomes, as have the peropsins classes (which persist in deuterostomes and some lophotrochozoa perhaps because without ciliary opsins there is no need for a retinal isomerase regeneration cycle). However it raises the question whether some neuroanatomical structures have also been lost.
The paired Drosophila retinas have 850 ommatidia each housing eight photoreceptors of three types; the paired cephalopharyngeal Bolwig organs have 12 photoreceptor cells of two types. Oddly, during metamorphosis to eyelet, the outer Bolwig cells die while inner cells switch gene expression from Rh6 to RH5.
Two of the Drosophila opsins have peak sensitivity in the ultraviolet (RH5 RH7) consistent with their K90 lysine and shorter motif, two sister opsins peak (RH3 RH4) in the blue and the rest (R6,(RH1,RH2)) at longer visible wavelengths. Opsins have been assigned to the four known photoreceptor structures as follows:
- RH5 RH6 Bolwig organ (larva) in founder and periferal cells, resp.
- RH6 Hofbauer-Buchner eyelet (adult founder cell remnants of Bolwig organ)
- RH2 ocellus (adult)
- RH1 R1-R6 periferal photoreceptors of ommatidia (adult eye)
- RH3 RH4 R7 photoreceptor of ommatidia (adult eye)
- RH5 RH6 R8 photoreceptor of ommatidia (adult eye)
- RH3 dorsal R7 R8 polaralization receptors (adult eye)
Note RH7 is missing from the list. This orphan opsin has no tissue-labelled transcripts at GenBank as of June 2009. It does not occur in any of the known photoreceptors, suggesting the repertoire of adult brain ultrastructures is still incomplete. Some authors have questioned whether RH7 is a 'real' opsin (because the third cytoplasmic loop CL3 is non-standard).
However it still retains the DRY motif, the Schiff base lysine and many other characteristic residues and opsin motifs. Its peak sensitivity would lie in the UV because of the well-conserved K90 motif, which is conserved in all 12 drosophilid genomes. The upstream PAX6 promoter RCSI site still matches the consensus sequence, TAATYCGATTA even though the first coding exon is anomalously lengthened and very prone to internal indels.
Other arthropod melanopsins also have unusual cytoplasmic third loops, which has predictive implications for Galpha signalling partner. This Galpha web tool allows studying the effects of replacing cytoplasmic loops or tail of RH7 with those of its nearest match, the UV-tuned RH5. This would not affect transmembrane structure or extracellular loops but might alter coevolved relations on the cytoplasmic face.
RH7 is exceedingly conserved (except in the amino terminus) in the other 11 drosophilids with sequenced genomes, ruling out both processed and unprocessed pseudogenes. Its two introns bear no relation in position and phase to those in any other drosophila opsins. The carboxy terminus is surprisingly conserved despite earlier indels. Remarkably for such a conserved gene, it is completely isolated phylogenetically. No other genomic insect or GenBank transcript provides even an ortholog candidate. This cannot plausibly reflect horizonal gene transfer (from what animal?) but cannot reflect an ancient gene duplication either.
Consequently the first order of business is to work up the species tree with targeted sequencing to pinpoint the evolutionary origin of RH7 -- Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha; Eremoneura; Cyclorrhapha; Schizophora; Acalyptratae; Ephydroidea; Drosophilidae; Drosophila melanogaster group. Because the gene is missing in dipteran and coleopteran genomes (mosquitoes, bee, flour beetle), that search can be be restricted. It seems too diverged from other opsins to have originated just in a few tens of millions of years of evolution represented by drosophilids (but perhaps not too much in terms of generations).
Second, it should be noted that a 2002 whole-proteome quantitative transcription project did in fact uncover RH7 transcripts (as displayed at the UCSC GeneSorter). Here peak expression, as normalized to egg-to-adult total RH7 transcripts, occured in 76-hour mesomorphs. Total expression was highest in 5-day adult females. Improved all-gene experiments in 2008-09 ruled out RH7 expression in pupae but verified expression in adult male and female heads at equal levels. These transcripts are not yet correlated with any anatomical structure. Despite arrays of the full set of 13,000 coding genes, a Drosophila brain expression atlas has never gotten off the ground -- each gene must be inefficiently studied in a one-off manner.
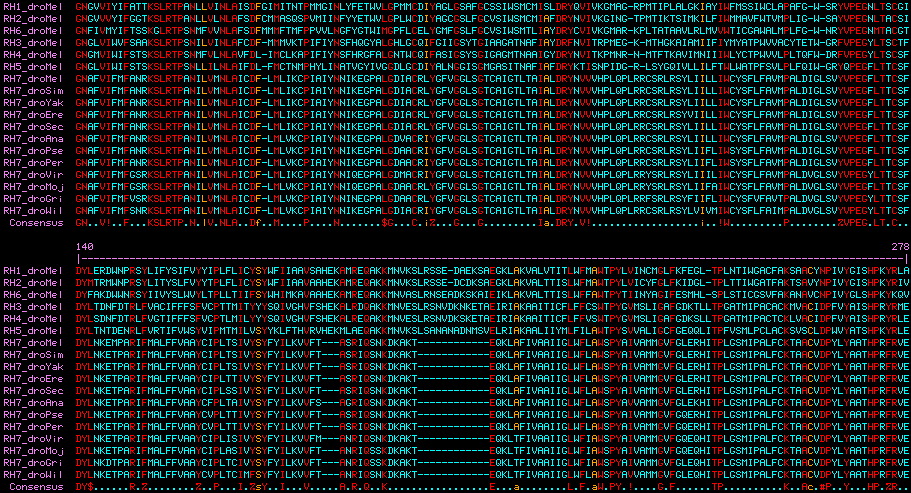
Gene, name, coding exons, introns present, chr location: RH1 (CG4550-RA) 5 chr3R 15,712,948 shares two introns with RH6 and one with RH2, similar SKA* termini RH2 (CG16740-RA) 4 chr3R 14,725,942 RH6 (CG5192-RB) 3 chr3R 11,309,650 RH3 (CG10888-RA) 1 chr3R 15,907,472 possible retrogene of RH5 RH4 (CG9668-RA) 2 chr3L 16,850,872 possible retrogene of RH5 with later intercolated genes RH5 (CG5279-RA) 3 chr2L 12,009,111 two ancestral introns (also Apis, Daphnia; first also Aplysia, Platynereis and Homo) RH7 (CG5638-RA) 3 chr3L 12,162,941 two novel introns, anomalous first exon
RH7 appears not involved in Drosophila circadian photoreception systems, which are mediated by the blue sensitive pterin-flavoprotein cryptochrome CRY (not homologous to opsins) in clock neurons and by opsins RH1, RH5 and RH6 in photoreceptors.
Curiously CRY is also implicated in magnetic field perception based anisotropic hyperfine coupling between unpaired electron and nuclear spins ([1, 2, 3). RH7 is not plausibly involved in this either because it is new whereas magnetosensing is old and widespread. In eery analogy to ciliary opsins, drosophilids but not butterflies have lost the close paralog to mammalian CRY.
>RH1_droMel Drosophila melanogaster (fruitfly) CG4550-RA
0 ME 00 SFAVAAAQLGPHFAPLSNGSVVDKVTPDMAHLISPYWNQFPAMDPIWAKILTAYMIMIGMISWCGNGVVIYIFATTKSLRTPANLLVINLAISDFGIMITNTPMMGINLYFETWVLGPMMCDIYAGLGSAFGCSSIWSMCMISLDRYQVIVKGMAGRPMTIPLALGKIAYIWFMSSIWCLAPAFGWSR 2
1 YVPEGNLTSCGIDYLERDWNPRSYLIFYSIFVYYIPLFLICYSYWFIIA 0
0 AVSAHEKAMREQAKKMNVKSLRSSEDAEKSAEGKLAKVALVTITLWFMAWTPYLVINCMGLFKFEGLTPLNTIWGACFAKSAACYNPIVYGIS 2
1 HPKYRLALKEKCPCCVFGKVDDGKSSDAQSQATASEAESKA* 0
>RH6_droMel Drosophila melanogaster (fruitfly) CG5192-RB gross genomic misassembly exon1
0 MASLHPPSFAYMRDGRNLSLAESVPAEIMHMVDPYWYQWPPLEPMWFGIIGFVIAILGTMSLAGNFIVMYIFTSSKGLRTPSNMFVVNLAFSDFMMMFTMFPPVVLNGFYGTWIMGPFLCELYGMFGSLFGCVSIWSMTLIAYDRYCVIVKGMARKPLTATAAVLRLMVVWTICGAWALM
PLFGWNRYVPEGNMTACGTDYFAKDWWNRSYIIVYSLWVYLTPLLTIIFSYWHIMK 0
0 AVAAHEKAMREQAKKMNVASLRNSEADKSKAIEIKLAKVALTTISLWFFAWTPYTIINYAGIFESMHLSPLSTICGSVFAKANAVCNPIVYGLS 2
1 HPKYKQVLREKMPCLACGKDDLTSDSRTQATAEISESQA* 0
>RH2_droMel Drosophila melanogaster (fruitfly) CG16740-RA
0 MERSHLPETPFDLAHSGPRFQAQSSGNGSVLDN 0
0 VLPDMAHLVNPYWSRFAPMDPMMSKILGLFTLAIMIISCCGNGVVVYIFGGTKSLRTPANLLVLNLAFSDFCMMASQSPVMIINFYYETWVLGPLWCDIYAGCGSLFGCVSIWSMCMIAFDRYNVIVKGINGTPMTIKTSIMKILFIWMMA
VFWTVMPLIGWSAYVPEGNLTACSIDYMTRMWNPRSYLITYSLFVYYTPLFLICYSYWFIIAAVAAHEKAMREQAKKMNVKSLRSSEDCDKSAEGKLAKVALTTISLWFMAWTPYLVICYFGLFKIDGLTPLTTIWGATFAKTSAVYNPIVYGIS 2
1 HPKYRIVLKEK 00 CPMCVFGNTDEPKPDAPASDTETTSEADSKA* 0
>RH3_droMel Drosophila melanogaster (fruitfly) CG10888-RA single exon
0 MESGNVSSSLFGNVSTALRPEARLSAETRLLGWNVPPEELRHIPEHWLTYPEPPESMNYLLGTLYIFFTLMSMLGNGLVIWVFSAAKSLRTPSNILVINLAFCDFMMMVKTPIFIYNSFH
QGYALGHLGCQIFGIIGSYTGIAAGATNAFIAYDRFNVITRPMEGKMTHGKAIAMIIFIYMYATPWVVACYTETWGRFVPEGYLTSCTFDYLTDNFDTRLFVACIFFFSFVCPTTMITYY
YSQIVGHVFSHEKALRDQAKKMNVESLRSNVDKNKETAEIRIAKAAITICFLFFCSWTPYGVMSLIGAFGDKTLLTPGATMIPACACKMVACIDPFVYAISHPRYRMELQKRCPWLALNEKAPESSAVASTSTTQEPQQTTAA* 0
>RH4_droMel Drosophila melanogaster (fruitfly) CG9668-RA two exons w large intron (no RM but intercolated genes)
0 MEPLCNASEPPLRPEARSSGNGDLQFLGWNVPPDQIQYIPEHWLTQLEPPASMHYMLGVFYIFLFCASTVGNGMVIWIFST
SKSLRTPSNMFVLNLAVFDLIMCLKAPIFIYNSFHRGFALGNTWCQIFASIGSYSGIGAGMTNAAIGYDRYNVITKPMNRNMTFTKAVIMNIIIWLYCTPWVVLPLTQFWDRFVP 1
2 EGYLTSCSFDYLSDNFDTRLFVGTIFFFSFVCPTLMILYYYSQIVGHVFSHEKALREQAKKMNVESLRSNVDKSKETAEIRIAKAAITICFLFFVSWTPYGVMSLI
GAFGDKSLLTPGATMIPACTCKLVACIDPFVYAISHPRYRLELQKRCPWLGVNEKSGEISSAQSTTTQEQQQTTAA* 0
>RH5_droMel Drosophila melanogaster (fruitfly) CG5279-RA two small introns also seen in Apis, Daphnia; first in Aplysia, Platynereis and Homo
0 MHINGPSGPQAYVNDSLGDGSVFPMGHGYPAEYQHMVHAHWRGFREAPIYYHAGFYIAFIVLMLSSIFGNGLVIWIFST 2
1 SKSLRTPSNLLILNLAIFDLFMCTNMPHYLINATVGYIVGGDLGCDIYALNGGISGMGASITNAFIAFDRYKTISNPIDGRLSYGQIVLLILFTWLWATPFSVLPLFQIWGRYQP 1
2 EGFLTTCSFDYLTNTDENRLFVRTIFVWSYVIPMTMILVSYYKLFTHVRVHEKMLAEQAKKMNVKSLSANANADNMSVELRIAKAALIIYMLFILAWTPYSVVALI
GCFGEQQLITPFVSMLPCLACKSVSCLDPWVYATSHPKYRLELERRLPWLGIREKHATSGTSGGQESVASVSGDTLALSVQN*
>RH7_droMel Drosophila melanogaster (fruitfly) CG5638-RA long N-terminal has M comp genomics support, EC074058 CO302368, 3 novel exons
0 MEAIIMTTLPNLTTDAGDSSFWLTGALSLSEMLANSSHSHSTGSTTSTAGSSATESSAVNVGKDHDKHVNDSVSTGLS 2
1 NYSNYPSYIHYRDKYDLSYIAKVNPFWLQFEPPKSSTFLIMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDI 1
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKEMPARIFMALFFVAAYCIPLTSIVYSYFYILKVVFTASRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGEGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNSISSVMEQSKF* 0
>RH7_droSim Drosophila simulans (fruitfly) chr3L:11530420 11532815
0 MEAIIMTTLPNLTTDAGDSSFWLTGALSLSEMLANSSHGHSTGSTTSSAGSSATESSAVNVGKDHDKHVNDSVSTGLS 2
1 NYSNYPSYIHYRDKYDLSYIAKVNPFWLQFEPPKSSTFLVMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDI 1
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLTSIVYSYFYILKVVFTASRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGEGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNSVSSVMEQSKF* 0
>RH7_droSec Drosophila sechellia (fruitfly) super_0:4344247 4346640
0 MEAIIMTTLPNLTTDAGDSSFWLTGALSLSEMLANSSHGHSTGSTASSAGSSATESSAVNVGKDHGKHVNDSVSTGLS 2
1 NYSNYPSYIHYRDKYDLSYIAKVNPFWLQFEPPKSSTFLVMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDI 1
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLSSIVYSYFYILKVVFTASRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGEGGICDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNSVSSVMEQSKF* 0
>RH7_droYak Drosophila yakuba (fruitfly) chr3L:12207286 12209654
0 MEAIIMTTLPALTTDAGDSSSFWLTGALSLSEMLANSSHGHSTGSTSSTAGSSATESSTVNVGKDHDVTKHVNDSVSTGLS 2
1 NYSNYPSYIHYRDKYDLSYIAKVNPFWLQFEPPKSSTFLVMAALYFLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDI 1
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLTSIVYSYFYILKVVFTASRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGDGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNSVSSVMEQSKF* 0
>RH7_droEre Drosophila erecta (fruitfly) scaffold_4784:12148112 12150459
0 MEAIIMTTLPTLTTDAGDSSFWLTGALSLSEMLANSSHGHSTGSTSSTAGSSATESATVNVGKDHDVAKHVNDSVSTGLS 2
1 NYSNYPSYIHYRDKYDLSYIAKVNPFWLQFEPPKSSTFLVMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDI 1
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYVIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLTTIVYSYFYILKVVFTASRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGDGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNSVSSVMEQSKF* 0
>RH7_droAna Drosophila ananassae (fruitfly) scaffold_13337:1483455 1485125+ frameshifted
0 MEAIILSTLPSLTTNASGSSSHWLTGALSLPEILANSSGSPNTSSADTGSGINLSARDADRHFNISTEAR 2
1 NYSYYPGYIHYRDKYDLSYIAKVNPFWLQFEPPHSSTFLAMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDV 1
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCFPLTAIVYSYFYILKVVFSAGRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAVVAMMGVFGLEKHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGILRRVSTTRSSYMTRSRSSFTHPAGRADGGTGRDHRMETYLMNNNLMMVPEETEENEEIVVVAEINNSVSSAIEQSKF* 0
>RH7_droPse Drosophila pseudoobscura (fruitfly) chrXR_group6:2491547 2493151
0 MEALMAALPTLTTEAAGSSLWLTSALSLSEMLANSSTSPNASLVAATTSSAAVATASTTSAAEAVGKVPDKHEVNDNVSTVLS 2
1 TSSSYPGYIHYRDKYDLSYIARVNPFWLQFEPPKSSTFYLMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDA 1
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIIFLIWSYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCVPLTTIVYSYFYILKVVFTASRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGQERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFFGRGVLRRVSTTRSSYMTRSRSSFNRRLRPTPDAEHRVESYLMNNNLMMVPEETEENEEIVVVAEFNNSSYSGMEQSKF* 0
>RH7_droPer Drosophila persimilis (fruitfly) super_9:783822 785423
0 MEALMAALPTLTTEAAGSSLWLTSALSLSEMLANSSTSPNASLVATTSSAAAATASTTSAAEAVGKVPDKHEVNDNVSTVLS 2
1 SYPGYIHYRDKYDLSYIARVNPFWLQFEPPKSSTFYLMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDA 1
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIIFLIWSYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCVPLTTIVYSYFYILKVVFTASRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGQERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFFGRGVLRRVSTTRSSYMTRSRSSFNRRLRPTPDAEHRVESYLMNNNLMMVPEETEENEEIVVVAEFNNSSYSGMEQSKF* 0
>RH7_droWil Drosophila willistoni (fruitfly) scaffold_180949:5140016 5141994+
0 MDMDMALDMNDAATTTSLWITSAALSLSEILVNTTSHVVTTSPASTSTVETTAVAAVTATGKVVHDDEKHHHHHHHHHQDEVNDNNVTTVLR 2
1 NFSSYPGYIHYRDKYDLSYIAKVNPFWLQFEPPRSSTFYIMAALYCLISVVGCIGNAFVIFMFSNRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDI 1
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLVIVMIWCYSFLFAIMPALDVGLSVYVPEGYLTTCSFDYLNKETPARIFMALFFVAAYCVPLTCIMFSYFYILKVVFTANRIQSNKDKAKTEQ
KLTFIVAAIIGLWFLAWSPYAVVAMMGVFGLEQHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLIYGRGVLRRVSTTRSSYITRSRSSFTRRLRTGSELDMRTEPYIMNNNLMMVPEETEENEEIVVVAEINNPSRCVSMHEHTSKF* 0
>RH7_droVir Drosophila virilis (fruitfly) scaffold_13049:6123835 6125790+
0 METIMSTFPTLTSDDGSLWITSALSEMLTSSSSNSSEAAQNATLVAAAAATTTTVAAAAAAAAANASTAATANVTKVHDKHSHAVNDSETDLR 2
1 CSAYPGYIHYRDKYDLDYIAKVNPFWLQFEPPGTSSFYIMAGLYCLISVVGCFGNAFVIFMFGSRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIQEGPALGDM 1
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRYSRLRSYLIIILIWCYSFLFAVMPALDVGLSVYVPEGYLTTCSFDYLNKETPARIFMALFFVAAYCIPLISIVYSYFYILKVVFMANRIQSNKDKAKTEQ
KLTFIVAAIIGLWFIAWSPYAIVAMMGVFGQEQHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLMYGRGALRRVSTTRSTYMTRSFTHRMRHTSGDGENRADPYTLNNNLMMVPEETEENDEIIVVAEINNSTSIAMEQSKF* 0
>RH7_droMoj Drosophila mojavensis (fruitfly) scaffold_6680:4445619 4446890+
0 METIMSTLPTLTADDGSLWITSALTELLASGANSSSGSSSVVADGTQNATFVAAATTTTTTVAAAAAAAAAAAVNASTATTANATKGHHKHPHGVNDSETDLR 2
1 LCSSYPGYIHYRDKYDLTYIAKVNPFWLQFEPPDTSTFYIMAALYCLISVVGCVGNAFVIFMFGSRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIQEGPALGDA 1
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRYSRLRSYLIIFAIWCYSFLFAVMPALDVGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLASIVYSYFYILKVVFTANRIQSSKDKAKTEQ
KLTFIVAAIIGLWFIAWSPYAIVAMMGVFGQEQHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLMYGRGVLRRVSTTRSSYMTRSRSSFTHRLRPSSGDCENRAEPYTLNNNLMMVPEETEENEEIIVVAEINNSISGVMEQSKF* 0
>RH7_droGri Drosophila grimshawi (fruitfly) scaffold_15110:6598464 6600409
0 METIMSTLPTLAADDGSQWLTSALSEVLASSDGRGAAQNATLAAATAVATATTAVNVSKVDDKHLHTVNDSDTDLT 2
1 RCSSYPGYIHYRDKYDLTYIAKVNPFWLQFEPPDTSTFYMMAGLYCLISVVGCFGNAFVIFMFVSRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNINEGPALGDA 1
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRFSRLRSYFIIFLIWCYSFVFAVTPALDVGLSVYVPEGYLTTCSFDYLNKDTPARIFMALFFVAAYCIPLTCIVYSYFYILKVVFTANRIQSSKDKAKTEQ
KLTFIVAAIIGLWFIAWSPYAIVAMMGVFGLEQHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMIFFGRGVLRRVSTTRSSYMTRSRSSFNHRVRSSSNEGDNRAESYKMNNNLMIVPEETDENEEIIVVAEINNSISIDMEQSKF* 0
1 130
RH7_droMel MEAIIMTTLPNLTTDAGDSS-FWLTGA-LSLSEMLANSS---------HSHSTG----STTSTAGSSATESSAV----NVGKDHD--KH---------VNDS-VSTGLSNYSNYPSYIHYRDKYDLSYIA
RH7_droSim MEAIIMTTLPNLTTDAGDSS-FWLTGA-LSLSEMLANSS---------HGHSTG----STTSSAGSSATESSAV----NVGKDHD--KH---------VNDS-VSTGLSNYSNYPSYIHYRDKYDLSYIA
RH7_droSec MEAIIMTTLPNLTTDAGDSS-FWLTGA-LSLSEMLANSS---------HGHSTG----STASSAGSSATESSAV----NVGKDHG--KH---------VNDS-VSTGLSNYSNYPSYIHYRDKYDLSYIA
RH7_droYak MEAIIMTTLPALTTDAGDSSSFWLTGA-LSLSEMLANSS---------HGHSTG----STSSTAGSSATESSTV----NVGKDHDVTKH---------VNDS-VSTGLSNYSNYPSYIHYRDKYDLSYIA
RH7_droEre MEAIIMTTLPTLTTDAGDSS-FWLTGA-LSLSEMLANSS---------HGHSTG----STSSTAGSSATESATV----NVGKDHDVAKH---------VNDS-VSTGLSNYSNYPSYIHYRDKYDLSYIA
RH7_droAna MEAIILSTLPSLTTNASGSSSHWLTGA-LSLPEILANSS---------GSPNTS----SADTGSGINLS-----------ARDAD--RH---------FN---ISTEARNYSYYPGYIHYRDKYDLSYIA
RH7_droPse MEAL-MAALPTLTTEAAGSS-LWLTSA-LSLSEMLANSS---------TSPNASLVAATTSSAAVATASTTSAA----EAVGKVPDKHE---------VNDN-VSTVLSTSSSYPGYIHYRDKYDLSYIA
RH7_droPer MEAL-MAALPTLTTEAAGSS-LWLTSA-LSLSEMLANSS---------TSPNASLVA-TTSSAAAATASTTSAA----EAVGKVPDKHE---------VNDN-VSTVLS---SYPGYIHYRDKYDLSYIA
RH7_droWil MDMDMALDMNDAATTTSLWITSAALSLSEILVNTT---------SHVVTTSPASTSTVETTAVAAVTATG----KVVHDDEKHHHHHHHHHQDEVNDNNVTTVLRNFSSYPGYIHYRDKYDLSYIA
RH7_droVir METI-MSTFPTLTSDDGS---LWITSALSEMLTSSSSNSS-------EAAQNATLVAAAAATTTTVAAAAAAAAA---NASTAATANVTKVHDKHSHAVNDS--ETDLR-CSAYPGYIHYRDKYDLDYIA
RH7_droMoj METI-MSTLPTLTADDGS---LWITSALTELLASGANSSSGSSSVVADGTQNATFVAAATTTTTTVAAAAAAAAAAAVNASTATTANATKGHHKHPHGVNDS--ETDLRLCSSYPGYIHYRDKYDLTYIA
RH7_droGri METI-MSTLPTLAADDGS---QWLTSALSEVLASSDGRG---------AAQNATLAAATAVATATTAV------------------NVSKVDDKHLHTVNDS--DTDLTRCSSYPGYIHYRDKYDLTYIA
Consensus me.......p.l..#.......W.T.A.......................................................................vNd....T.l...S.YP.YIHYRDKYDL.YIA
131 260
RH7_droMel KVNPFWLQFEPPKSSTFLIMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDIACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSR
RH7_droSim KVNPFWLQFEPPKSSTFLVMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDIACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSR
RH7_droSec KVNPFWLQFEPPKSSTFLVMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDIACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSR
RH7_droYak KVNPFWLQFEPPKSSTFLVMAALYFLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDIACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSR
RH7_droEre KVNPFWLQFEPPKSSTFLVMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDIACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSR
RH7_droAna KVNPFWLQFEPPHSSTFLAMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDVACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSR
RH7_droPse RVNPFWLQFEPPKSSTFYLMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDAACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSR
RH7_droPer RVNPFWLQFEPPKSSTFYLMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDAACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSR
RH7_droWil KVNPFWLQFEPPRSSTFYIMAALYCLISVVGCIGNAFVIFMFSNRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDIACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSR
RH7_droVir KVNPFWLQFEPPGTSSFYIMAGLYCLISVVGCFGNAFVIFMFGSRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIQEGPALGDMACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRYSR
RH7_droMoj KVNPFWLQFEPPDTSTFYIMAALYCLISVVGCVGNAFVIFMFGSRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIQEGPALGDAACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRYSR
RH7_droGri KVNPFWLQFEPPDTSTFYMMAGLYCLISVVGCFGNAFVIFMFVSRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNINEGPALGDAACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRFSR
Consensus kVNPFWLQFEPP..StF..MA.LYCLISVVGC.GNAFVIFMF..RKSLRTPANILVMNLAICDFLML!KCPIAIYNNI.EGPALGD.ACR.YGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRR.SR
261 390
RH7_droMel LRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKEMPARIFMALFFVAAYCIPLTSIVYSYFYILKVVFTASRIQSNKDKAKTEQKLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLER
RH7_droSim LRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLTSIVYSYFYILKVVFTASRIQSNKDKAKTEQKLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLER
RH7_droSec LRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLSSIVYSYFYILKVVFTASRIQSNKDKAKTEQKLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLER
RH7_droYak LRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLTSIVYSYFYILKVVFTASRIQSNKDKAKTEQKLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLER
RH7_droEre LRSYVIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLTTIVYSYFYILKVVFTASRIQSNKDKAKTEQKLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLER
RH7_droAna LRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCFPLTAIVYSYFYILKVVFSAGRIQSNKDKAKTEQKLAFIVAAIIGLWFLAWSPYAVVAMMGVFGLEK
RH7_droPse LRSYLIIFLIWSYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCVPLTTIVYSYFYILKVVFTASRIQSNKDKAKTEQKLAFIVAAIIGLWFLAWSPYAIVAMMGVFGQER
RH7_droPer LRSYLIIFLIWSYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCVPLTTIVYSYFYILKVVFTASRIQSNKDKAKTEQKLAFIVAAIIGLWFLAWSPYAIVAMMGVFGQER
RH7_droWil LRSYLVIVMIWCYSFLFAIMPALDVGLSVYVPEGYLTTCSFDYLNKETPARIFMALFFVAAYCVPLTCIMFSYFYILKVVFTANRIQSNKDKAKTEQKLTFIVAAIIGLWFLAWSPYAVVAMMGVFGLEQ
RH7_droVir LRSYLIIILIWCYSFLFAVMPALDVGLSVYVPEGYLTTCSFDYLNKETPARIFMALFFVAAYCIPLISIVYSYFYILKVVFMANRIQSNKDKAKTEQKLTFIVAAIIGLWFIAWSPYAIVAMMGVFGQEQ
RH7_droMoj LRSYLIIFAIWCYSFLFAVMPALDVGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLASIVYSYFYILKVVFTANRIQSSKDKAKTEQKLTFIVAAIIGLWFIAWSPYAIVAMMGVFGQEQ
RH7_droGri LRSYFIIFLIWCYSFVFAVTPALDVGLSVYVPEGYLTTCSFDYLNKDTPARIFMALFFVAAYCIPLTCIVYSYFYILKVVFTANRIQSSKDKAKTEQKLTFIVAAIIGLWFIAWSPYAIVAMMGVFGLEQ
Consensus LRSYl!I..IWcYSFlFA!mPALD!GLSVYVPEG%LTTCSFDYLNK#TPARIFMALFFVAAYC.PL..Iv%SYFYILKVVF.A.RIQS.KDKAKTEQKL.FIVAAIIGLWF.AWSPYA!VAMMGVFG.E.
391 517
RH7_droMel HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGEGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNS---ISSVMEQSKF
RH7_droSim HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGEGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNS---VSSVMEQSKF
RH7_droSec HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGEGGICDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNS---VSSVMEQSKF
RH7_droYak HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGDGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNS---VSSVMEQSKF
RH7_droEre HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGDGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNS---VSSVMEQSKF
RH7_droAna HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGILRRVSTTRSSYMTRSRSSFTHPAGRADGGTGR--DHRMETYLMNNNLMMVPEETEENEEIVVVAEINNS---VSSAIEQSKF
RH7_droPse HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFFGRGVLRRVSTTRSSYMTRSRSSFNRRLRPTPDAE-----HRVESYLMNNNLMMVPEETEENEEIVVVAEFNNS---SYSGMEQSKF
RH7_droPer HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFFGRGVLRRVSTTRSSYMTRSRSSFNRRLRPTPDAE-----HRVESYLMNNNLMMVPEETEENEEIVVVAEFNNS---SYSGMEQSKF
RH7_droWil HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLIYGRGVLRRVSTTRSSYITRSRSSFTRRLRTGSEL-----DMRTEPYIMNNNLMMVPEETEENEEIVVVAEINNPSRCVSMHEHTSKF
RH7_droVir HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLMYGRGALRRVSTTRSTYMTRS---FTHRMRHTSGD----GENRADPYTLNNNLMMVPEETEENDEIIVVAEINNS---TSIAMEQSKF
RH7_droMoj HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLMYGRGVLRRVSTTRSSYMTRSRSSFTHRLRPSSGD----CENRAEPYTLNNNLMMVPEETEENEEIIVVAEINNS---ISGVMEQSKF
RH7_droGri HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMIFFGRGVLRRVSTTRSSYMTRSRSSFNHRVRSSSNE----GDNRAESYKMNNNLMIVPEETDENEEIIVVAEINNS---ISIDMEQSKF
Consensus HITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMl.%GRG.LRRVSTTRSsYmTRSrssF..r.r............R.#.Y.$NNNLMmVPEET#EN#EI!VVAEiNNs....s...eqSKF