Genome Browser Session Gallery: Difference between revisions
From genomewiki
Jump to navigationJump to search
(New page: {| class="wikitable" border="0"|- |- | == Tier 1 Cell Lines == 400px|thumb|left|Tier 1 cell lines [http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&...) |
(Updated hgwdev/genome-test links to .gi, one link broken) |
||
| (13 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
|- | |- | ||
| | | | ||
== Tier 1 Cell Lines == | == ENCODE Tier 1 Cell Lines == | ||
[[Image:EncodeKent1.gif| | [[Image:EncodeKent1.gif|800px|thumb|none|Tier 1 cell lines]] | ||
[http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent1 Launch Browser] | [http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent1 Launch Browser] | ||
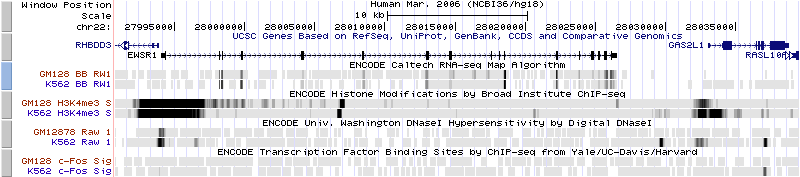
Basic ENCODE data on the Tier 1 cell lines GM128 | Basic ENCODE data on the Tier 1 cell lines GM128 (a lymphoid | ||
cell line | cell line) and K562 (a myeloid line). This track collection includes: | ||
which shows the level of expression of the gene EWSR1 | * RNA-seq, which shows the level of expression of the gene EWSR1 | ||
histone mark | * the histone mark H3K4Me3, which is associated with promoters | ||
hypersensitivity, which is associated with regulatory regions in general | * DNAseI hypersensitivity, which is associated with regulatory regions in general | ||
* ChIP-seq, which shows levels of occupancy by the transcription factor c-Fos | |||
(Contributed by Jim Kent.) | |||
| Line 19: | Line 21: | ||
== Premature Stop in Reference Assembly == | == Premature Stop in Reference Assembly == | ||
[[Image:EncodeKent2.gif| | [[Image:EncodeKent2.gif|800px|thumb|none|Premature stop in reference assembly]] | ||
[http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent2 Launch Browser] | [http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent2 Launch Browser] | ||
| Line 33: | Line 35: | ||
== Multiple Histone Marks == | == Multiple Histone Marks == | ||
[[Image:EncodeKent3.gif| | [[Image:EncodeKent3.gif|800px|thumb|none|Multiple histone marks.]] | ||
[http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent3 Launch Browser] | [http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent3 Launch Browser] | ||
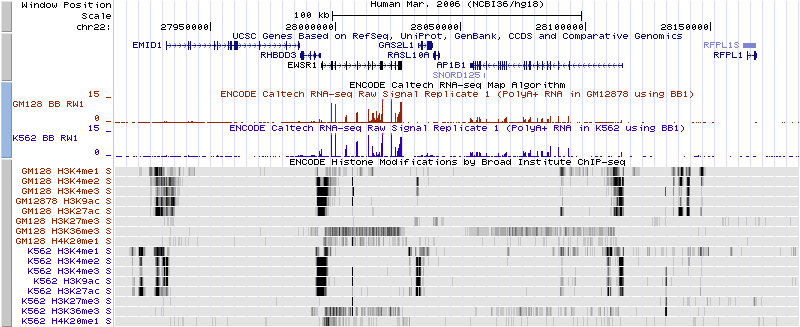
Histone ChIP-seq data on two ENCODE Tier 1 cell lines. | |||
H3K4me1 is associated with enhancers and (to an extent) with promoters. | H3K4me1 is associated with enhancers and (to an extent) with promoters. | ||
H3K4me3 is associated with promoters, and H3K36me3 is associated with transcribed regions. | H3K4me3 is associated with promoters, and H3K36me3 is associated with transcribed regions. | ||
| Line 46: | Line 48: | ||
|- | |- | ||
| | | | ||
<!-- Commented out til reg tracks are released | |||
== Session from Ross Hardison == | |||
[[Image:EncodeHardison1.gif|800px|thumb|none]] | |||
BROKEN | |||
[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Rosshardison&hgS_otherUserSessionName=8q24_9kb_KateR Launch Browser] -- dang. this session no longer exists. | |||
(Contributed by Ross Hardison.) | |||
|- | |||
| | |||
--> | |||
== Template == | == Template == | ||
[[Image:image.jpg| | [[Image:image.jpg|600px|thumb|none|text below image]] | ||
[http://urlToSession Launch Browser] | [http://urlToSession Launch Browser] | ||
| Line 56: | Line 70: | ||
Contributed by . | |||
|} | |} | ||
Latest revision as of 23:54, 31 August 2018
ENCODE Tier 1 Cell LinesBasic ENCODE data on the Tier 1 cell lines GM128 (a lymphoid cell line) and K562 (a myeloid line). This track collection includes:
(Contributed by Jim Kent.)
|
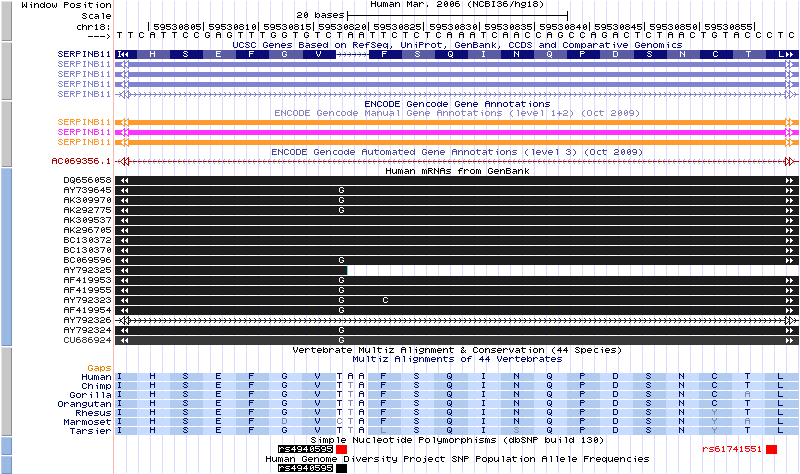
Premature Stop in Reference AssemblyA premature stop codon that is found in the reference genome and about half of people of European descent. The UCSC Genes track is forced to skip the codon.
|
Multiple Histone MarksHistone ChIP-seq data on two ENCODE Tier 1 cell lines. H3K4me1 is associated with enhancers and (to an extent) with promoters. H3K4me3 is associated with promoters, and H3K36me3 is associated with transcribed regions.
|
|
TemplateDescription of scenario.
|