Opsin evolution: LWS PhyloSNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) No edit summary |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
'''See also:''' [[Opsin_evolution|Curated Sequences]] | [[Opsin_evolution:_Encephalopsin_gene_loss|Encephalopsins]] | [[Opsin_evolution:_Melanopsin_gene_loss|Melanopsins]] | [[Opsin_evolution:_Neuropsin_phyloSNPs|Neuropsin]] | [[Opsin_evolution:_Peropsin_phyloSNPs|Peropsin]] | [[Opsin_evolution:_RGR_phyloSNPs|RGR phyloSNPs]] | [[Opsin_evolution:_update_blog|Update Blog]] | |||
== Comparative Genomics of LWS opsins == | == Comparative Genomics of LWS opsins == | ||
The LWS (long wave sensitive) cone opsin gene is especially favorable for comparative genomics because numerous pre-existing studies on specific species and the happenstance of NISC targeting have greatly expanded the depth of phylogenetic coverage available from the 50 genomic projects and accidental EST coverage, bringing the total coverage close to 100 species of vertebrates. This affords the opportunity to examine the robustness of phyloSNPs deduced just from genomic projects to a doubling of sampling density. | The LWS (long wave sensitive) cone opsin gene is especially favorable for comparative genomics because numerous pre-existing studies on specific species and the happenstance of NISC targeting have greatly expanded the depth of phylogenetic coverage available from the 50 genomic projects and accidental EST coverage, bringing the total coverage close to 100 species of vertebrates. This affords the opportunity to examine the robustness of phyloSNPs deduced just from genomic projects to a doubling of sampling density. | ||
It is quickly apparent however that GenBank sequences cannot be used at face value; they are error-ridden, with more than a handful of entries having spurious (disabling) indels and highly implausible regional bursts of radical amino acid substitutions. In some cases, multiple independent sequences allow confident correction of error, though it must be noted that transcript and genomic projects rarely reference the same individual animal (meaning some polymorphism can be expected). Because no central physical depository for dna exists, it may not be possible to ever | It is quickly apparent however that GenBank sequences cannot be used at face value; they are error-ridden, with more than a handful of entries having spurious (disabling) indels and highly implausible regional bursts of radical amino acid substitutions. In some cases, multiple independent sequences allow confident correction of error, though it must be noted that transcript and genomic projects rarely reference the same individual animal (meaning some polymorphism can be expected). Because no central physical depository for dna exists, it may not be possible to ever re-sequence many of the anomalous entries. | ||
GenBank entries for LWS genes display a bizarre range of confused descriptors (green opsin, red opsin, red-green opsin, red-sensitive pigment, red-green sensitive, medium-wave-sensitive, long-wave-sensitive, iodopsin, LWSA, M/L, MW, LW, M/LWS, KFH-R , cp-L, AYU-R, LWS_QUEm5_L06, color blindness deutan, color blindness protan). Less than half the entries including standard terms such LWS or OPN1LW that would allow simple database retrieval using the registered gene name. GenBank has been [http://www.sciencemag.org/cgi/content/short/319/5870/1616a roundly criticized] by 256 signatories for its refusal to allow third-party corrections. | GenBank entries for LWS genes display a bizarre range of confused descriptors (green opsin, red opsin, red-green opsin, red-sensitive pigment, red-green sensitive, medium-wave-sensitive, long-wave-sensitive, iodopsin, LWSA, M/L, MW, LW, M/LWS, KFH-R , cp-L, AYU-R, LWS_QUEm5_L06, color blindness deutan, color blindness protan). Less than half the entries including standard terms such LWS or OPN1LW that would allow simple database retrieval using the registered gene name. GenBank has been [http://www.sciencemag.org/cgi/content/short/319/5870/1616a roundly criticized] by 256 signatories for its refusal to allow third-party corrections. | ||
| Line 15: | Line 17: | ||
Then, using the [http://mbe.oxfordjournals.org/cgi/reprint/15/5/560 "five sites rule"] of Yokoyama, as subsequently [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=1460773&blobtype=pdf amended] and [http://www.genetics.org/cgi/content/full/158/4/1697 refined], the peak adsorption can be rather reliably predicted using S180A, H197Y, Y277F, T285A, and A308S as shifting toward blue by 7, 28, 8, 15, and 27 nm from a base level of P560. These are not quite additive as the double change S180A/H197Y shifts lambda max by 11 nm. | Then, using the [http://mbe.oxfordjournals.org/cgi/reprint/15/5/560 "five sites rule"] of Yokoyama, as subsequently [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=1460773&blobtype=pdf amended] and [http://www.genetics.org/cgi/content/full/158/4/1697 refined], the peak adsorption can be rather reliably predicted using S180A, H197Y, Y277F, T285A, and A308S as shifting toward blue by 7, 28, 8, 15, and 27 nm from a base level of P560. These are not quite additive as the double change S180A/H197Y shifts lambda max by 11 nm. | ||
This could be revisited today in view of a vastly expanded data set and the [http://genome.cshlp.org/content/early/2009/02/04/gr.084509.108.full.pdf+html excellent discussion of tuning residues] in chondrichthyes LWS. Here elephant shark exhibits a lineage-specific duplication of LWS. These have | This could be revisited today in view of a vastly expanded data set and the [http://genome.cshlp.org/content/early/2009/02/04/gr.084509.108.full.pdf+html excellent discussion of tuning residues] in chondrichthyes LWS. Here elephant shark exhibits a lineage-specific duplication of LWS. These have lambda max of 499 nm and 548 nm after reconstitution with 11-cis-retinal, with with wavelength shift driven by the H181/K184 chloride site (not operative in LWS1) but primarily A292S (with serine inactivating). | ||
This serine substitution, presumably with the same effect, is also seen at position 292 in mouse, rat, rabbit, pika, dolphin, whales, and hedgehog. Glires (rodents and lagomorphs) all have tyrosine at position 181 in place of histidine, though still K184. | This serine substitution, presumably with the same effect, is also seen at position 292 in mouse, rat, rabbit, pika, dolphin, whales, and hedgehog. Glires (rodents and lagomorphs) all have tyrosine at position 181 in place of histidine, though still K184. | ||
Using Yokoyama offsets, lambda max in the curated set of LWS genes below range from 505 in eriEur (shrew) to 560 in sciCar (squirrel). These tuning residues | Using Yokoyama offsets, lambda max in the curated set of LWS genes below range from 505 in eriEur (shrew) to 560 in sciCar (squirrel). These tuning residues potentially explain the highly restricted reduced alphabet at these positions (with an alternatives observed only at S180P in lampreys and two fish), despite the greatly expanded the species set. These 5 residues are shown in the alignment markup track below which are easily extracted for spreadsheet analysis without distraction from extraneous residues. Spectra for all lamprey cone opsins other than LWS (P560 predicted) have been [http://www.fasebj.org/cgi/content/full/21/11/2713 experimentally measured.] | ||
It would be necessary to comb the literature for all available experimentally determined wavelengths prior to re-parameterization. So many species have been directly measured that predictions for residual species scarcely add value. Here it must be noted that chromophores such as vitamin A2 are used in some species (perhaps at some mixing level) and impact actual lambda max whereas reconstituted proteins can be measured with reference to vitamin A1. Ancestral spectra need recalculation at each divergence node with today's much higher species sampling density and experimental testing in full length ancestral protein | It would be necessary to comb the literature for all available experimentally determined wavelengths prior to re-parameterization. So many species have been directly measured that predictions for residual species scarcely add value. Here it must be noted that chromophores such as vitamin A2 are used in some species (perhaps at some mixing level) and impact actual lambda max whereas reconstituted proteins can be measured with reference to vitamin A1. Ancestral spectra need recalculation at each divergence node with today's much higher species sampling density and experimental testing in full length ancestral protein reconstructions. | ||
The diagram shows the location of residues tuning lambda max in the five imaging opsin gene classes. It could be improved by considering the full reduced alphabets seen in high density sampling and instituting a cutoff for significant change as ultimately nearly every substitution at nearly every central position will have some effect however small on the chromophore environment. Further, a shift in one opsin should not be considered in isolation but rather in the context of the overall visionary system together with the photic environment experienced by the animal. | The diagram shows the location of residues tuning lambda max in the five imaging opsin gene classes. It could be improved by considering the full reduced alphabets seen in high density sampling and instituting a cutoff for significant change as ultimately nearly every substitution at nearly every central position will have some effect however small on the chromophore environment. Further, a shift in one opsin should not be considered in isolation but rather in the context of the overall visionary system together with the photic environment experienced by the animal. | ||
| Line 33: | Line 35: | ||
== Locus control region (LCR) between SWS2 and LWS == | == Locus control region (LCR) between SWS2 and LWS == | ||
SWS2 and LWS occur as a tandem pair ancestrally, as implied by a conserved association from teleost fish through amniotes and monotremes (note SWS2 has been deleted in the stem to marsupials and placentals). A cis-acting locus control regulatory region, analogous to that of the beta hemoglobin cluster, sits between the two genes and regulates both. It was not affected by the SWS2 deletion event; indeed, as LWS duplicated downstream in old world primates, the LWS:LCR region extended its reach to regulate this gene as well. Color imaging opsin genes must be regulated so that they are expressed in the retina only and only in cone cells there often in a | SWS2 and LWS occur as a tandem pair ancestrally, as implied by a conserved association from teleost fish through amniotes and monotremes (note SWS2 has been deleted in the stem to marsupials and placentals). A cis-acting locus control regulatory region, analogous to that of the beta hemoglobin cluster, sits between the two genes and regulates both. It was not affected by the SWS2 deletion event; indeed, as LWS duplicated downstream in old world primates, the LWS:LCR region extended its reach to regulate this gene as well. Color imaging opsin genes must be regulated so that they are expressed in the retina only and only in cone cells -- and there often in a defined gradient pattern. | ||
[[Image:LwsLCR.png|left|]] | [[Image:LwsLCR.png|left|]] | ||
| Line 45: | Line 47: | ||
<br clear="all"> | <br clear="all"> | ||
The evidence is weaker that the two groups of LWS:LCR sequences are orthologous. Ordinary alignment is not convincing in part because the sequences are only 70 bp to begin with and the time of group divergence is 310 myr and more. However both sets of sequences are positioned similarly as recognized phastCons elements downstream of SWS2 and upstream of LWS. Experimental data in human, mouse, frog, and | The evidence is weaker that the two groups of LWS:LCR sequences are orthologous. Ordinary alignment is not convincing in part because the sequences are only 70 bp to begin with and the time of group divergence is 310 myr and more. However both sets of sequences are positioned similarly as recognized phastCons elements downstream of SWS2 and upstream of LWS. Experimental data in human, mouse, frog, and zebrafish support similar cis-acting roles. Further, the lizard comparative genomics track method recovers the platypus LWS:LCR sequence at the expected place relative to lizard, frog, and fish LWS:LCR elements. | ||
Thus it appears the LWS:LCR regulatory element arose prior to lamprey divergence and has retained its position upstream of LWS in all descendent clades, yet | Thus it appears the LWS:LCR regulatory element arose prior to lamprey divergence and has retained its position upstream of LWS in all descendent clades, yet significant evolutionary changes occurred in the earliest mammals (conceivably associated with Walls' postulated nocturnal era in conjunction with loss of SWS2 in therans). Additional sequences relevant to this transitional era (eg more early amniotes, echidna, more marsupials) may illuminate these changes but more likely insufficient extant species exist. | ||
LWS occupies a special basal position in the vertebrate imaging opsin gene tree -- a subsequent duplication cascade gave rise to the other opsins. This raises the question of whether the ancestral LWS:LCR regulatory element also propagated to these other opsins which, except for SWS2, are at unrelated chromosomal positions. Since these opsins retained the fine details of exon structure, the duplication events were not retropositioning and reintronation, ie were some form of block duplication that may well have brought the LWS:LCR regulatory element along because otherwise the new gene could not be plausibly targeted to retinal cone cells prior to acquiring disabling mutations. | LWS occupies a special basal position in the vertebrate imaging opsin gene tree -- a subsequent duplication cascade gave rise to the other opsins. This raises the question of whether the ancestral LWS:LCR regulatory element also propagated to these other opsins which, except for SWS2, are at unrelated chromosomal positions. Since these opsins retained the fine details of exon structure, the duplication events were not retropositioning and reintronation, ie were some form of block duplication that may well have brought the LWS:LCR regulatory element along because otherwise the new gene could not be plausibly targeted to retinal cone cells prior to acquiring disabling mutations. | ||
| Line 53: | Line 55: | ||
Note that the opsin gene tree (as deduced independently in dozens of publications) directly conflicts with the sistering tree topology expected from supposed whole genome duplications. That is, the chain of duplication is LWS -> SWS1 -> SWS2 -> RHO2 -> RHO1. Why then is SWS2 rather than SWS1 adjacent to LWS? | Note that the opsin gene tree (as deduced independently in dozens of publications) directly conflicts with the sistering tree topology expected from supposed whole genome duplications. That is, the chain of duplication is LWS -> SWS1 -> SWS2 -> RHO2 -> RHO1. Why then is SWS2 rather than SWS1 adjacent to LWS? | ||
A key experimental observation here is tandem duplications of LWS still fall under the control of the LWS:LCR regulatory element even though that element itself may not have been duplicated. This provides a temporal window of retained functionality during which the | A key experimental observation here is tandem duplications of LWS still fall under the control of the LWS:LCR regulatory element even though that element itself may not have been duplicated. This provides a temporal window of retained functionality during which the spectral properties of the new gene can diverge with selective advantage, for example to peak sensitivity at shorter wavelengths. We know such events have been fixed numerous times (the SWS2 event pre-lamprey, the [http://genome.cshlp.org/content/early/2009/02/04/gr.084509.108.full.pdf+html LWS duplication in elephantfish] which is very likely tandem, the MLS event in primates, in howler monkeys, and in various teleost fish). | ||
One speculative scenario: the block tandomly duplicated giving rise to an ancestral SWS1 with its own (soon-to-diverge) upstream copy of regulatory element called here SWS1:LCR. After protein divergence (that explains the closer relation of SWS2 to SWS1 than to LWS), the SWS1 gene but not its regulatory element duplicated tandomly downstream to give SWS1:LCR SWS1 SWS2 LWS:LCR LWS. A later chromosomal rearrangement broke off the now self-sufficient SWS1 from the cluster. This scenario then repeated itself, but with a twist as SWS2 alone duplicated internally, gave rise to upstream RHO2:LCR and RHO2. RHO2 later gave rise to RHO1 which is targeted still to retina but to rod cells. This event may have taken place separately post-lamprey giving confusing RHO2 comparisons. This scenario distinctively predicts regulatory elements upstream for LWS and SWS1 but downstream/shared for SWS2 and upstream of RHO2 and RHO1 coding. | One speculative scenario: the block tandomly duplicated giving rise to an ancestral SWS1 with its own (soon-to-diverge) upstream copy of regulatory element called here SWS1:LCR. After protein divergence (that explains the closer relation of SWS2 to SWS1 than to LWS), the SWS1 gene but not its regulatory element duplicated tandomly downstream to give SWS1:LCR SWS1 SWS2 LWS:LCR LWS. A later chromosomal rearrangement broke off the now self-sufficient SWS1 from the cluster. This scenario then repeated itself, but with a twist as SWS2 alone duplicated internally, gave rise to upstream RHO2:LCR and RHO2. RHO2 later gave rise to RHO1 which is targeted still to retina but to rod cells. This event may have taken place separately post-lamprey giving confusing RHO2 comparisons. This scenario distinctively predicts regulatory elements upstream for LWS and SWS1 but downstream/shared for SWS2 and upstream of RHO2 and RHO1 coding. | ||
| Line 82: | Line 84: | ||
[[Image:SWS_CRX.png|left|]] | [[Image:SWS_CRX.png|left|]] | ||
Similarly, while upstream regulatory regions of SWS1 have been [http://mend.endojournals.org/cgi/content/full/20/8/1728 experimentally studied], including a putative CRX binding site (LCR), there is no basis for | Similarly, while upstream regulatory regions of SWS1 have been [http://mend.endojournals.org/cgi/content/full/20/8/1728 experimentally studied], including a putative CRX binding site (LCR), there is no basis for inferring homology to LWS control regions. Indeed sites of best sequence conservation, in the form of phastCons features from the 28-species alignment, correlate most imperfectly with the short CRX and orphan nuclear receptor RORB binding motifs (consensus CTAATC and AGGTCA respectively). The 3 unassigned peaks of conservation could correspond to as yet undescribed transcription regulatory proteins. | ||
It is highly implausible that a small block translocation of a coding gene would evolve a de novo regulatory element from adjacent junk dna (even the small CRX and RX motifs together with 39 bp dimer spacing are a formidable barrier) or land near a pre-existing appropriate regulatory element (that of an adjacent unrelated gene targeted to retina cones at appropriate developmental window) even though element sharing can occur. | It is highly implausible that a small block translocation of a coding gene would evolve a de novo regulatory element from adjacent junk dna (even the small CRX and RX motifs together with 39 bp dimer spacing are a formidable barrier) or land near a pre-existing appropriate regulatory element (that of an adjacent unrelated gene targeted to retina cones at appropriate developmental window) even though element sharing can occur. | ||
| Line 408: | Line 410: | ||
93.letJap CSL..........................D.......V.......FL...V.I..........HS..Q......T.....RD.S......I..YI......T....Y.......A....T.....Y........................M............V....R.......NS.I... | 93.letJap CSL..........................D.......V.......FL...V.I..........HS..Q......T.....RD.S......I..YI......T....Y.......A....T.....Y........................M............V....R.......NS.I... | ||
94.geoAus C...........................TD.......V...I...FI..AL.II.........HT..Q......T.....RD.S......IF.YI......T............A....A.....Y..........I.............M............V..SAR.......NS..... | 94.geoAus C...........................TD.......V...I...FI..AL.II.........HT..Q......T.....RD.S......IF.YI......T............A....A.....Y..........I.............M............V..SAR.......NS..... | ||
</pre> | |||
=== The marsupial insert in exon 1 | |||
<pre> | |||
For example, this novel insertion in the first exon of LWS is a pretty good one for uniting the Didelphimorphia to the exclusion of the other marsupials: | |||
LWS_homSap MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS | |||
LWS_gorGor MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS | |||
LWS_ponPyg MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS | |||
LWS_panTro MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS | |||
LWS_nomLeu MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS | |||
LWS_papHam MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS | |||
LWS_macMul MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS | |||
LWS_macFas MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS | |||
LWS_cal539 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFIYTNSNS | |||
LWS_cal553 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFIYTNSNS | |||
LWS_ceb530 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFTYTNSNS | |||
LWS_cal561 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFTYTNSNS | |||
LWS_ceb560 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFTYTNSNS | |||
LWS_ceb545 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFTYTNSNS | |||
LWS_ate560 MAQQWSLQRLAGRHPQDSYE---DSTQSSVFTYTNNNA | |||
LWS_ate552 MAQQWSLQRLAGRHPQDSYE---DSTQSSVFTYTNNNA | |||
LWS_otoGar MAQQWGPQRLTGRQPQDTHE---DSTQGSIFTYTNSNT | |||
LWS_otoCra MAQQWGPQRLTGRQPQDTHE---DSTQGSIFTYTNSNT | |||
LWS_micMur MAQQWGPHRFAGGQPQDSHE---DSTQASIFTYTNSNA | |||
LWS_tupBel MAQRWGPHKLAGGQPQDSYE---DSTQSSIFTYTNTNS | |||
LWS_musMus MAQ------LTGEQTLDHYE---DSTHASIFTYTNSNS | |||
LWS_ratNor MAQ------LTGEQTLDHYE---DSTQASIFTYTNSNS | |||
LWS_nanEhr MAQQWAPQRLAGGQTQDSYE---DSTQASLFTYTNSNS | |||
LWS_speTri MAQRWDPQRLAGGQPQDSHE---DSTQSSIFTYTNSNA | |||
LWS_oryCun MTQPWGPQMLAGGQPPESHE---DSTQASIFTYTNSNS | |||
LWS_cavPor MAQRWGPHALSGVQAQDAYE---DSTQASLFTYTNSNN | |||
LWS_canFam MTQRWGPQRLAGGQPQAGLE---ESTQASIFTYTNSNA | |||
LWS_felCat MTQRWGPQRLAGGQPHAGLE---DSTRASIFTYTNSNA | |||
LWS_bosTau MAHAWGPQRLAGGQPQANFE---ESTQGSIFTYTNSNS | |||
LWS_susScr MAQQWGPRRLAGGQPQASFE---DSTQGSIFTYTNANS | |||
LWS_equCab MAQRWGPQKLAGGQPQAGFE---DSTQASIFTYTNNNA | |||
LWS_phoVit MAQTWGPQRLADGRPQPGYE---DSTQASIFTYTNSNA | |||
LWS_myoLuc MAQRWGPQRLAGGQLQASFE---DSTLASIFTYTNSNA | |||
LWS_pteVam MAQSWVPQRLAEGQPQASFE---ESTQASIFIYTNSNV | |||
LWS_turTru MAQQWGPQRFAGGQPQTSFE---DSTQGSVFTYTNSNS | |||
LWS_gloMel MAQQWGPQRFAGGQPQTSFE---DSTQGSVFTYTNSNS | |||
LWS_delDel MAQTWGPQRFAGGQPQTSFE---DSTQGSVFTYTNSNS | |||
LWS_loxAfr MAQQWGPHRLTGARLQDASE---DSTQASIFVYTNTNT | |||
LWS_proCap MAEPWGTHRLTGRQLPDASE---DSTQASIFVYTNSNA | |||
LWS_echTel MAQRWGAHRLTGGQLQDTYE---GSTRTSIFVYTNSTS | |||
LWS_triMan MAQQWGTHRLTGGQPQDAYE---DSTQASIFVYTNTNT | |||
LWS_monDom MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNN Didelphimorphia | |||
LWS_didAur MTQAWDPVGFLARRRDENEDDHDDTTRASLFVYTNSNN Didelphimorphia | |||
LWS_tarRos MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodontia | |||
LWS_macEug MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodontia | |||
LWS_smiCra MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Dasyuromorphia | |||
LWS_myrFas MTQAWDPAGFLAWRREENE----ETTRASLFTYTNSNN Dasyuromorphia | |||
LWS_setBra MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodonti | |||
LWS_cerCon MTQAWDPAGFLAWQEDENE----ETTRASLFVYTNSNN Diprotodontia | |||
LWS_isoObe MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Peramelemorphia | |||
LWS_ornAna MTPAWNSGVYAARRRFEDEE---DTTRTSVFVYTNSNN | |||
LWS_tacAcu MTQAWDPAGFLAWRRDENEE---TTRASLFVYTNSNNT | |||
LWS_galGal MAAWEAA--FAARRRHEEE----DTTRDSVFTYTNSNN | |||
LWS_taeGut MATWDGAV-FAARRRHDDE----DTTRDSIFTYTNSNN | |||
LWS_colLiv M---DG---FAAARRRHEDE---DTTRDSVFTYTNSNN | |||
LWS_anoCar MAEAWDVAVFAARRRNDED----DTTRDSLFTYTNSNN | |||
LWS_gecGec MTEAWNVAVFAARRSRDDD----DTTRGSVFTYTNTNN | |||
LWS_pheMad MTEAWNVAVFAARRSRDDDD---DTTRGSVFTYTDSNN | |||
LWS_cynPyr MAYSWNSGAYAARRRYDDE----DTTRSSVFVYTNSNN | |||
LWS_ambTig MAHSWNSGAYAARRRYDDE----DTTRSSIFTYTNSNN | |||
LWS_xenTro MASHWNEAVFAARRRNDDD----DTTRSSVFTYTNSNN | |||
LWS_xenLae MASQLNEAIFAARRRNDDD----DTTRSSVFTYTNSNN | |||
LWS_neoFor MAEPWD-AVLAARRRHQDE----ETTRSTIFVYTNSNN | |||
LWS_danRer MAEHWGDAIYAARRKGD------ETTREAMFTYTNSNN | |||
LWS_cypCar MAEQWGDAIFAARRRGD------ETTRETMFVYTNSNN | |||
LWS_pleAlt MTDEWGNAVFAARRRNE------DTTRESSFTYTNSNN | |||
LWS_tetNig MAEEWGKQSFAARRYHE------DSTRGSAFVYTNSNH | |||
LWS_takRub MAEEWGKQSFAARRYHE------DTTRGSAFVYTNSNH | |||
LWS_ophVen MAEEWGKQSFAARRYHE------DSTRGSAFAYTNSNN | |||
LWS_oryLat MAEQWGKQVFAARRQNE------DTTRGSAFTYTNSNH | |||
LWS_lucGoo MAEQWEKQAFAARRYNE------ETTRGSVFTYTNSNH | |||
LWS_gasAcu MAEEWGKQAFAARRYNE------DTTRGSMFVYTNSNN | |||
LWS_pseMax MAEDWGKPAFAARRYHE------DTTRGSAFMYTNSNH | |||
LWS_poeRet MAEEWGKQVFAARR-HE------DTTRGAAFTYTNSNH | |||
LWS_salSal MAERWGSAAYAARRQNQ------DTTRESSFTFTNSNN | |||
LWS1_calMi MTQSWELVAPAARRGFKYD----EPTHSGIFVYTNSNQ | |||
LWS2_calMi MAEPRGSVAFAARR-WNDH----EGTTVGEFTYTNSNS | |||
LWS_sciCar MAQRWDPQRLAGGQPQDSHE---DSTQSSIFTYTNSNA | |||
LWS_eriEur MAQREGPQRLAGVQPQDGLE---SSTLGSTFTYSGSNS | |||
LWS_petMar MTASWQGAMFAARRRQDDE----DTTMESLFRYTNENN | |||
LWS_letJap MTASWHGAVFAARRRNDDE----DTTKDSIFRYTNENN | |||
LWS_geoAus MAQSWERAMFAARRRQDE-----DTTKGDLFRYTNENN | |||
</pre> | </pre> | ||
| Line 957: | Line 1,048: | ||
0 FRNCILQLFGKKVDDSSELSSTSKMEASSASSVSPA* 0 | 0 FRNCILQLFGKKVDDSSELSSTSKMEASSASSVSPA* 0 | ||
>LWS_monDom Monodelphis domesticus (opossum) | >LWS_monDom Monodelphis domesticus (opossum) Didelphimorphia) VNED insert | ||
0 | 0 MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNNTR 1 | ||
2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 | 2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 | ||
2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTAPPLFGWSR 2 | 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTAPPLFGWSR 2 | ||
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYVQVWLAIRA 0 | 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYVQVWLAIRA 0 | ||
0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYSFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYSFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | ||
0 FRTCILQLFGKKVDDGSEVSSTSKTEGSSVSSVAPA* 0 | |||
>LWS_didAur Didelphis aurita (big-eared opossum) Didelphimorphia ENED insert | |||
0 MTQAWDPVGFLARRRDENEDDHDDTTRASLFVYTNSNNTR 1 | |||
2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 | |||
2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTSPPLFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSSDLGVQSYMIVLMATCCIFPLSIILLCYIQVWLAIRA 0 | |||
0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA* 0 | |||
>LWS_tarRos Tarsipes rostratus (honey possum) Diprotodontia | |||
0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 | |||
2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | |||
2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAIWTSPPLFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGNSDPGIQSYMIVLMSTCCILPLSIILLCYVQVWRAIRA 2 | |||
0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 | 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 | ||
>LWS_macEug Macropus eugenii (wallaby) | >LWS_macEug Macropus eugenii (wallaby) Diprotodontia | ||
0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 | 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 | ||
2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | 2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | ||
| Line 973: | Line 1,080: | ||
0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 | 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 | ||
>LWS_setBra Setonix brachyurus (quokka) Diprotodontia | |||
>LWS_setBra Setonix brachyurus (quokka) | |||
0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 | 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 | ||
2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETMIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | 2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETMIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | ||
| Line 989: | Line 1,088: | ||
0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVPA* 0 | 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVPA* 0 | ||
>LWS_cerCon Cercartetus concinnus ( | >LWS_cerCon Cercartetus concinnus (pygmy possum) Diprotodontia | ||
0 MTQAWDPAGFLAWQEDENEETTRASLFVYTNSNNTK 1 | 0 MTQAWDPAGFLAWQEDENEETTRASLFVYTNSNNTK 1 | ||
2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAIADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | 2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAIADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | ||
| Line 997: | Line 1,096: | ||
0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* | 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* | ||
> | >LWS_smiCra Sminthopsis crassicaudata (dunnart) Dasyuromorphia | ||
0 | 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 | ||
2 | 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | ||
2 | 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 | ||
1 | 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIILCYIQVWLAIRA 0 | ||
0 | 0 VAKQQKESESTQKAEKEVSRMVMVMILAFCFCWGPYALFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | ||
0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 | |||
>LWS_myrFas Myrmecobius fasciatus (numbat) Dasyuromorphia | |||
0 MTQAWDPAGFLAWRREENEETTRASLFTYTNSNNTK 1 | |||
2 GPFEGPNYHIAPRWVYNLTSFWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | |||
2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSVILLCYIQVWLAIRA 0 | |||
0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 | 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 | ||
>LWS_isoObe Isoodon obesulus (bandicoot) | >LWS_isoObe Isoodon obesulus (bandicoot) Peramelemorphia | ||
0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 | 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 | ||
2 GPFEGPNYHIAPRWVYNLTSFWMFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | 2 GPFEGPNYHIAPRWVYNLTSFWMFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | ||
| Line 1,012: | Line 1,119: | ||
0 VAKQQKDSESTQKAEKEVSRMVVVMIRAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | 0 VAKQQKDSESTQKAEKEVSRMVVVMIRAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | ||
0 FRTCILQLFGKKVDDGSEVSGTSRTEVSSVSSAPA* 0 | 0 FRTCILQLFGKKVDDGSEVSGTSRTEVSSVSSAPA* 0 | ||
>LWS_ornAna Ornithorhynchus anatinus (platypus) full | >LWS_ornAna Ornithorhynchus anatinus (platypus) full | ||
| Line 1,279: | Line 1,378: | ||
</pre> | </pre> | ||
'''See also:''' [[Opsin_evolution|Curated Sequences]] | [[Opsin_evolution:_Encephalopsin_gene_loss|Encephalopsins]] | [[Opsin_evolution:_Melanopsin_gene_loss|Melanopsins]] | [[Opsin_evolution:_Neuropsin_phyloSNPs|Neuropsin]] | [[Opsin_evolution:_Peropsin_phyloSNPs|Peropsin]] | [[Opsin_evolution:_RGR_phyloSNPs|RGR phyloSNPs]] | [[Opsin_evolution:_update_blog|Update Blog]] | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 11:22, 23 March 2010
See also: Curated Sequences | Encephalopsins | Melanopsins | Neuropsin | Peropsin | RGR phyloSNPs | Update Blog
Comparative Genomics of LWS opsins
The LWS (long wave sensitive) cone opsin gene is especially favorable for comparative genomics because numerous pre-existing studies on specific species and the happenstance of NISC targeting have greatly expanded the depth of phylogenetic coverage available from the 50 genomic projects and accidental EST coverage, bringing the total coverage close to 100 species of vertebrates. This affords the opportunity to examine the robustness of phyloSNPs deduced just from genomic projects to a doubling of sampling density.
It is quickly apparent however that GenBank sequences cannot be used at face value; they are error-ridden, with more than a handful of entries having spurious (disabling) indels and highly implausible regional bursts of radical amino acid substitutions. In some cases, multiple independent sequences allow confident correction of error, though it must be noted that transcript and genomic projects rarely reference the same individual animal (meaning some polymorphism can be expected). Because no central physical depository for dna exists, it may not be possible to ever re-sequence many of the anomalous entries.
GenBank entries for LWS genes display a bizarre range of confused descriptors (green opsin, red opsin, red-green opsin, red-sensitive pigment, red-green sensitive, medium-wave-sensitive, long-wave-sensitive, iodopsin, LWSA, M/L, MW, LW, M/LWS, KFH-R , cp-L, AYU-R, LWS_QUEm5_L06, color blindness deutan, color blindness protan). Less than half the entries including standard terms such LWS or OPN1LW that would allow simple database retrieval using the registered gene name. GenBank has been roundly criticized by 256 signatories for its refusal to allow third-party corrections.
Less than 5% of the entries mention the peak adsorption (lambda max) even when that was experimentally determined and indeed the whole purpose for sequencing. In some cases, the abstract provides the critical data; in other cases, access to proprietary text is needed; in still others, no article was ever published referencing the submitted sequence.
Consequently, the GenBank annotation -- if not the sequence itself -- is often worthless. It is necessary to begin from scratch with a fixed 'type specimen' and expand that out to a core set of LWS genes with syntenically verified orthology. That was done here earlier in building the original opsin classifier. Each putative new LWS ortholog must then unequivocally cluster with the core set distinctly from all other (cone) opsin gene family members. There are various complexities at the level of the individual species that are not the focus here, such as triallelic species. LWS has experienced a fair number of lineage-specific tandem and whole-genome duplications (notably in fish), pseudogenization, and gene losses (new here: Xenarthra).
Then, using the "five sites rule" of Yokoyama, as subsequently amended and refined, the peak adsorption can be rather reliably predicted using S180A, H197Y, Y277F, T285A, and A308S as shifting toward blue by 7, 28, 8, 15, and 27 nm from a base level of P560. These are not quite additive as the double change S180A/H197Y shifts lambda max by 11 nm.
This could be revisited today in view of a vastly expanded data set and the excellent discussion of tuning residues in chondrichthyes LWS. Here elephant shark exhibits a lineage-specific duplication of LWS. These have lambda max of 499 nm and 548 nm after reconstitution with 11-cis-retinal, with with wavelength shift driven by the H181/K184 chloride site (not operative in LWS1) but primarily A292S (with serine inactivating).
This serine substitution, presumably with the same effect, is also seen at position 292 in mouse, rat, rabbit, pika, dolphin, whales, and hedgehog. Glires (rodents and lagomorphs) all have tyrosine at position 181 in place of histidine, though still K184.
Using Yokoyama offsets, lambda max in the curated set of LWS genes below range from 505 in eriEur (shrew) to 560 in sciCar (squirrel). These tuning residues potentially explain the highly restricted reduced alphabet at these positions (with an alternatives observed only at S180P in lampreys and two fish), despite the greatly expanded the species set. These 5 residues are shown in the alignment markup track below which are easily extracted for spreadsheet analysis without distraction from extraneous residues. Spectra for all lamprey cone opsins other than LWS (P560 predicted) have been experimentally measured.
It would be necessary to comb the literature for all available experimentally determined wavelengths prior to re-parameterization. So many species have been directly measured that predictions for residual species scarcely add value. Here it must be noted that chromophores such as vitamin A2 are used in some species (perhaps at some mixing level) and impact actual lambda max whereas reconstituted proteins can be measured with reference to vitamin A1. Ancestral spectra need recalculation at each divergence node with today's much higher species sampling density and experimental testing in full length ancestral protein reconstructions.
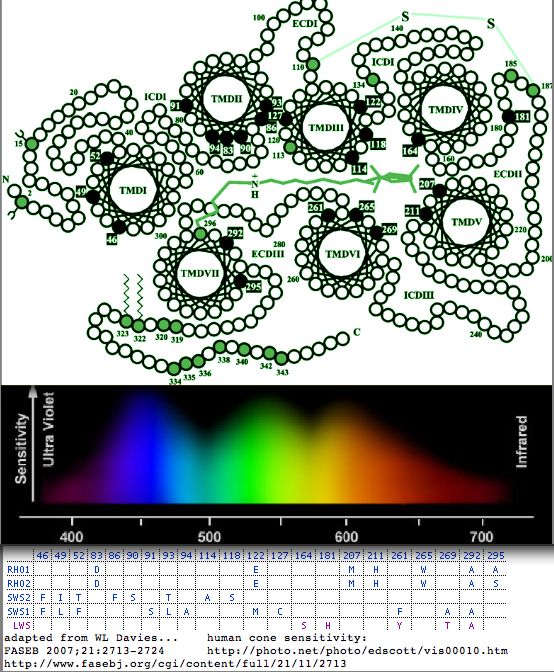
The diagram shows the location of residues tuning lambda max in the five imaging opsin gene classes. It could be improved by considering the full reduced alphabets seen in high density sampling and instituting a cutoff for significant change as ultimately nearly every substitution at nearly every central position will have some effect however small on the chromophore environment. Further, a shift in one opsin should not be considered in isolation but rather in the context of the overall visionary system together with the photic environment experienced by the animal.
More ambitiously, all opsins might be considered for a comprehensive prediction program. The paralogous reduced alphabets at the five sites can vary quite considerably in non-cone opsins, for example in peropsins, neuropsins, and rgropsins. Here it is not so clear whether these five residues are still the principal tuning residues (as might be expected from structural conservation) nor what the functional significance of lambda max shifts might be in non-imaging opsins.
In the LWS alignment below, it can be seen that variations the five tuning positions are phylogenetically incoherent (not phyloSNPs). Note though however coherence within Glires (rodents + rabbits) of the tyrosine at position 197. It's not clear what the specific adaptive value of this 28 nm blueshift might be; perhaps further sampling would dissipate the apparent coherence.
Locus control region (LCR) between SWS2 and LWS
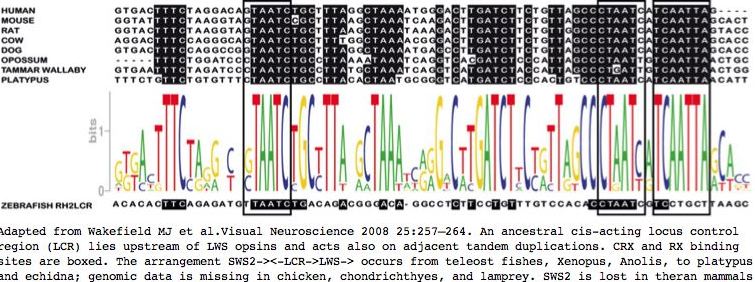
SWS2 and LWS occur as a tandem pair ancestrally, as implied by a conserved association from teleost fish through amniotes and monotremes (note SWS2 has been deleted in the stem to marsupials and placentals). A cis-acting locus control regulatory region, analogous to that of the beta hemoglobin cluster, sits between the two genes and regulates both. It was not affected by the SWS2 deletion event; indeed, as LWS duplicated downstream in old world primates, the LWS:LCR region extended its reach to regulate this gene as well. Color imaging opsin genes must be regulated so that they are expressed in the retina only and only in cone cells -- and there often in a defined gradient pattern.
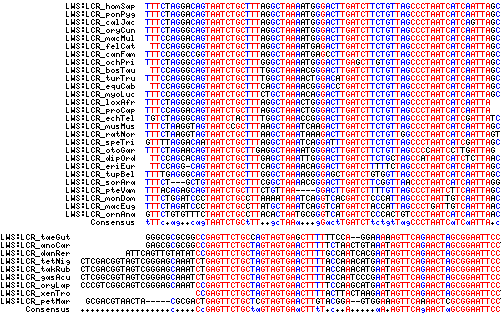
Here, the LWS:LCR region is recovered from 35 species ranging from human to lamprey, expanding on recent work of MJ Wakefield et al. The group of mammalian LWS:LCR sequences are clearly orthologous, based on experimental evidence, high percentage identity, syntenic location and orientation with respect to LWS strand, uniqueness as Blat matches within whole genomes, and genomic alignment provided in the UCSC 28-species alignment (where LWS:LCR is a significant phastCons element). By the same criteria, the sequences from teleost fish, frog, and lizard are also orthologous.
Lamprey also is 80% identical to the consensus sequence from this group and retains conserved nucleotides, but the genome assembly has gaps in this region and the anticipated intermediate position in lamprey SWS2-LWS cannot be validated. This could be better pursued in the Geotria genome which has a full complement of opsins.
The evidence is weaker that the two groups of LWS:LCR sequences are orthologous. Ordinary alignment is not convincing in part because the sequences are only 70 bp to begin with and the time of group divergence is 310 myr and more. However both sets of sequences are positioned similarly as recognized phastCons elements downstream of SWS2 and upstream of LWS. Experimental data in human, mouse, frog, and zebrafish support similar cis-acting roles. Further, the lizard comparative genomics track method recovers the platypus LWS:LCR sequence at the expected place relative to lizard, frog, and fish LWS:LCR elements.
Thus it appears the LWS:LCR regulatory element arose prior to lamprey divergence and has retained its position upstream of LWS in all descendent clades, yet significant evolutionary changes occurred in the earliest mammals (conceivably associated with Walls' postulated nocturnal era in conjunction with loss of SWS2 in therans). Additional sequences relevant to this transitional era (eg more early amniotes, echidna, more marsupials) may illuminate these changes but more likely insufficient extant species exist.
LWS occupies a special basal position in the vertebrate imaging opsin gene tree -- a subsequent duplication cascade gave rise to the other opsins. This raises the question of whether the ancestral LWS:LCR regulatory element also propagated to these other opsins which, except for SWS2, are at unrelated chromosomal positions. Since these opsins retained the fine details of exon structure, the duplication events were not retropositioning and reintronation, ie were some form of block duplication that may well have brought the LWS:LCR regulatory element along because otherwise the new gene could not be plausibly targeted to retinal cone cells prior to acquiring disabling mutations.
Note that the opsin gene tree (as deduced independently in dozens of publications) directly conflicts with the sistering tree topology expected from supposed whole genome duplications. That is, the chain of duplication is LWS -> SWS1 -> SWS2 -> RHO2 -> RHO1. Why then is SWS2 rather than SWS1 adjacent to LWS?
A key experimental observation here is tandem duplications of LWS still fall under the control of the LWS:LCR regulatory element even though that element itself may not have been duplicated. This provides a temporal window of retained functionality during which the spectral properties of the new gene can diverge with selective advantage, for example to peak sensitivity at shorter wavelengths. We know such events have been fixed numerous times (the SWS2 event pre-lamprey, the LWS duplication in elephantfish which is very likely tandem, the MLS event in primates, in howler monkeys, and in various teleost fish).
One speculative scenario: the block tandomly duplicated giving rise to an ancestral SWS1 with its own (soon-to-diverge) upstream copy of regulatory element called here SWS1:LCR. After protein divergence (that explains the closer relation of SWS2 to SWS1 than to LWS), the SWS1 gene but not its regulatory element duplicated tandomly downstream to give SWS1:LCR SWS1 SWS2 LWS:LCR LWS. A later chromosomal rearrangement broke off the now self-sufficient SWS1 from the cluster. This scenario then repeated itself, but with a twist as SWS2 alone duplicated internally, gave rise to upstream RHO2:LCR and RHO2. RHO2 later gave rise to RHO1 which is targeted still to retina but to rod cells. This event may have taken place separately post-lamprey giving confusing RHO2 comparisons. This scenario distinctively predicts regulatory elements upstream for LWS and SWS1 but downstream/shared for SWS2 and upstream of RHO2 and RHO1 coding.
Proposed sequence of events giving rise to vertebrate visual opsins explaining opsin gene tree and chromosomal proximities.
Key issues: SWS2 closer to LWS in gene tree yet SWS1 syntenic with LWS and using its control region
Status of chrX locus Other loci Comment
LCR-LWS original gene targeted to cone
LCR-LWS LCR-LWS first duplication: LWS and regulatory element
LCR-SWS1 LCR-LWS SWS1 attributes emerge
LCR-SWS1 LCR-SWS1 LCR-LWS second duplication: just SWS1 and its LCR
LCR-SWS1 LCR-SWS2 LCR-LWS SWS2 attributes emerge in middle gene
LCR-SWS2 LCR-LWS LCR-SWS1 translocation of SWS1 and its LCR to an autosomal chr
LCR-SWS2 SWS2 LCR-LWS LCR-SWS1 third duplication: SWS2 but not its control region
LCR-RH02 SWS2 LCR-LWS LCR-SWS1 RHO2 attributes emerge, SWS2 transcribed by LWS control region
SWS2 LCR-LWS LCR-SWS1 LCR-RH02 translocation of RHO2 and its LCR to new autosomal chr
SWS2 LCR-LWS LCR-SWS1 LCR-RH02 LCR-RH01 fourth duplication:RHO2, emergence and translocation of RHO1
LCR-LWS LCR-SWS1 LCR-RH02 LCR-RH01 SWS2 lost in theran mammals, SWS1 lost in monotremes
LCR-LWS MWS LCR-SWS1 LCR-RH02 LCR-RH01 fifth duplication: LWS in OW primates but not control region
In this scenario, the regulatory elements diverge paralogously from their common ancestor just like the corresponding opsin genes. Note the history of descent differs slightly from that of the gene tree in that SWS2:LCR descends with RHO2 as SWS2 has no regulatory element of its own. Testing this would require finding these paralogous regulatory elements and establishing comparative genomics nesting.
This raises the question of where the regulatory element first arose in progenitors to LWS, namely pinopsin (a gene restricted to frog through bird expressed in pineal), VAOP (fish through bird), parapinopsin (tunicates to bird), and parietopsin. None of these genes were retained in mammal, so the bioinformatics of finding homologs to LCR are unfavorable. As the retina arose prior to lamprey divergence, gene duplicates destined to provide imaging opsins needed to be targeted to emerging cone cells, even as parent genes retained expression in pineal. Thus the regulatory circuits here must already have been well-established 540 myr ago in the Cambrian.
A potential paralog RHO2:LCR has been studied experimentally in the zebrafish tetrameric tandem. Here an upstream RH2:LCR element controls the four downstream opsin genes consistent with the scenario non-vacuously in that regulatory elements can occur separated, upstream, or downstream of their targeted genes. Unfortunately, that element has no bioinformatically detectable homologous counterparts even in other species of fish with determined genomes; the opsin itself is lost in mammals.
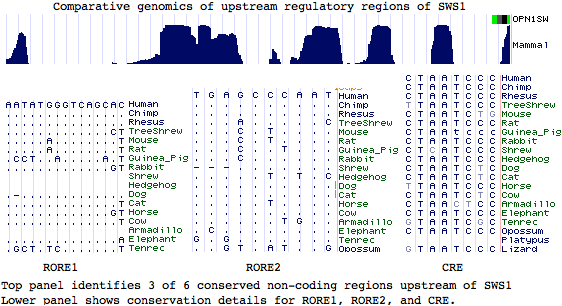
Similarly, while upstream regulatory regions of SWS1 have been experimentally studied, including a putative CRX binding site (LCR), there is no basis for inferring homology to LWS control regions. Indeed sites of best sequence conservation, in the form of phastCons features from the 28-species alignment, correlate most imperfectly with the short CRX and orphan nuclear receptor RORB binding motifs (consensus CTAATC and AGGTCA respectively). The 3 unassigned peaks of conservation could correspond to as yet undescribed transcription regulatory proteins.
It is highly implausible that a small block translocation of a coding gene would evolve a de novo regulatory element from adjacent junk dna (even the small CRX and RX motifs together with 39 bp dimer spacing are a formidable barrier) or land near a pre-existing appropriate regulatory element (that of an adjacent unrelated gene targeted to retina cones at appropriate developmental window) even though element sharing can occur.
35 LCR control region sequences and 2 consensus sequences >LWS:LCR_homSap Homo sapiens (human) tttctaggacagtaatctgctttaggctaaaatgggacttgatcttctgttagccctaatcatcaattagc >LWS:LCR_ponPyg Pongo pygmaeus (orang_sumatran) tttctaggacagtaatctgctttaggctaaaatgggacttgatcttctgttagccctaatcatcaattagc >LWS:LCR_macMul Macaca mulatta (rhesus) tttccaggacagtaatctgctttaggctaaaatgggacttgatcttctgttagccctaatcatcaattagc >LWS:LCR_calJac Callithrix jacchus (marmoset) tttctaggacagtaatctgctttaggctaaaatgggacttgatcttctgttagccctaatcatcaGttagc >LWS:LCR_otoGar Otolemur garnettii (bushbaby) tttctagaacagtaatctgcttgaggctaaaatgggacttgatcttctgttagccccaatccttgattag >LWS:LCR_tupBel Tupaia belangeri (tree_shrew) ttttgagggcagtaatctgctttgggctaaaaggggagctgatctgtggttagccctaatcatcaattagc >LWS:LCR_musMus Mus musculus (mouse) tttctaaggtagtaatccgctttaagctaaatcaagacttgatcttctgttagccctaatcatcaattagc >LWS:LCR_ratNor Rattus norvegicus (rat) tttctaaggtagtaatctgctttaagctaaataaagacttgatcttctgttggccctaatcatcaattagt >LWS:LCR_speTri Spermophilus tridecemlineatus (squirrel) gttttaggacaataatctgctttaggctaaatcaggatttgatcttctgttagccctaatcatcgattagc >LWS:LCR_dipOrd Dipodomys ordii (kangaroo_rat) ttccagcacagtaatctgctttgagctaaattgggacttgatcttctgctagccataatcatctcttaac >LWS:LCR_oryCun Oryctolagus cuniculus (rabbit) tttctagggcagtaatctgctttaggctaaaatgggacttgatcttctgttagccctaatcatcaattagc >LWS:LCR_ochPri Ochotona princeps (pika) tttctagggcagtaatctgctttgggctaaaatgggacttgagcttgtgttagccctaatcatcaattagc >LWS:LCR_canFam Canis familiaris (dog) tttccaggccggtaatctgctttaggctaaaatgagccttgatcttctgttagccctaatcatcaattagc >LWS:LCR_felCat Felis catus (cat) ttccagggcagtaatctgctttaggctaaaatgagacttgatcttctgttagccctaatcatcaattagc >LWS:LCR_bosTau Bos taurus (cow) ttccagggcagtaatctgcttttggctaaaacgggacttgatcttctgttagccctaatcatcaattagc >LWS:LCR_equCab Equus caballus (horse) ttccagggcagtaatctgctttcagctaaaacgggacctgatcttctgttagccctaatcatcaattagc >LWS:LCR_myoLuc Myotis lucifugus (microbat) tttccagggcagtaatctgctttctgctaaaacaggacttgatcttctgttagccctaatcatcaattagc >LWS:LCR_pteVam Pteropus vampyrus (macrobat) 4 bp del tttacagagcagtaatctgctttctgttaag gacttgatcttttgttagccctaatcttcaattagc >LWS:LCR_turTru Tursiops truncatus (dolphin) tttccagggcagtaatctgcttttggctaaaactggacatgatcttctgttagccctaatcatcaattagc >LWS:LCR_eriEur Erinaceus europaeus (hedgehog) ttcca ggcagtaatctgctttcagctaaaacaggacttgatctttttctagccctaatcatcaattagc >LWS:LCR_sorAra Sorex araneus (shrew) tttct gctgtaatctgctttcggctaaaacgggacttgatcttcggttaaccctaatcatcaattagg >LWS:LCR_loxAfr Loxodonta africana (elephant) tttctagggcagtaatctgctttaggctaaactgggacttgatcttctgttagccctaatcatcaatta >LWS:LCR_proCap Procavia capensis (hyrax) tttccaggacagtaatctgctttaggctaaactgggacttgatcttctgttagccctaatcatcaatta >LWS:LCR_echTel Echinops telfairi (tenrec) tgtctagggcagtaatctacttttggctaaaccgggacttgatcttctgttagccctaatcatcgattatc >LWS:LCR_monDom Monodelphis domestica (opossum) tttctggatccctaatctgccttaaaataaatcaggtcacgatctcccattagccctaattgtcaattaac >LWS:LCR_macEug Macropus eugenii (wallaby) tttctagatccctaatctgccttatgctaaatcaggtcatgatctaccattagccctgattgtcaattaac >LWS:LCR_ornAna Ornithorhynchus anatinus (platypus) gttctgtgtttctaatctgccttacactaatgcgggtcatgatctcccactgtccctaatcatcaattaac >LWS:LCR_mamCon mammalian consenus sequence tTTctaggacagTAATCTGCtTTaggcTAAaatggGactTGATCTtctgtTagCCCTAATCaTCaATTAgc >LWS:LCR_taeGut Taeniopygia guttata (finch) 86% identical; from anoCar probe gggcgcgcggccgagttctgccagtagtgagctttt ttccaggaaaaagttcagaactagcggaattcc >LWS:LCR_anoCar Anolis carolinensis (lizard) gagcgcgcggccgagttctgctagtagtgaactttttctaactgtaaatagttcagaactagcggaattcctgac >LWS:LCR_xenTro Xenopus tropicalis (frog) catg removed ccgagttctgctagtagtgaacttttacttactaa gaatagttcagaactagcggaattcctgacaagcac >LWS:LCR_danRer Danio rerio (zebrafish) attcagttgtatatccgagttctgctagtagtgaacttttgccaatcacgaatagttcagaactagcggaattccaggcatacgactgact >LWS:LCR_tetNig Tetraodon nigroviridis (pufferfish) ctcgacggt agtcgggagcaaatctgagttctgctagtagtgaacttttaccaaacatgaatagttcagaactagcggaattccagacctgctctcgaaa >LWS:LCR_takRub Takifugu rubripes (fugu) ctcgacggt agtcgggagcaaatctgagttctgctagtagtgaacttttaccaaacatgaatagttcagaactagcggaattccagacctgccctcgaaa >LWS:LCR_gasAcu Gasterosteus aculeatus (stickleback) ctcggcggtagtcgggagcaaatctgagttctgctagtagtgaacttttaccaatcccgaatagttcagaactagcggaattccagacctaccctcgaaa >LWS:LCR_oryLap Oryzias latipes (medaka) cccgtcggc agtcgggagcaaatccgagttctgctagtagtgaactttttccaagcatgaatagttcagaactagcggaattccagacctactctcgaaa >LWS:LCR_petMar Petromyzon marinus (lamprey) traces 1.2e-06 80% gcgacgtaactaCGCGACTGAGTTCTGCTCGTAGTGAACTTGTACGGAGTGGAAA GTTCAAAACTCGCGGAATTCCTGCCGGGCA >LWS:LCR_telCon fish-lizard-bird consenus sequence CcGAGTTCTGCtAGTAGTGAaCTTTTactaactatgAAtAGTTCAGAACTAGCGGAATTCCagac
PhyloSNPs in LWS opsins: alignment of 94 LWS vertebrate opsins
.position ...................................................................................................1.........1.........1.........1.........1.........1.........1.........1.........1..1 .position .........1.........2.........3.........4.........5.........6.........7.........8.........9.........0.........1.........2.........3.........4.........5.........6.........7.........8..8 .position 123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123 .....exon 111111111111111111111111111111111111122222222222222222222222222222222222222222222222222222222222222222222222222222222222222222222222222233333333333333333333333333333333333333333333333 ..markers .................................gly.......................................................................................diS.cI....................ERY...........................P-7. 01.homSap MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTRGPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLCGITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWAAVW 02.panTro ...................................................................................................................S....................................L.................I........S... 03.gorGor ....................................................................................................................................................................................... 04.ponPyg ................................................................T..................................................S.........................................................V...V..... 05.nomLeu ................................................................T..................................................S....................................L.................I........S... 06.macMul ...................................................................................................................S.........................................................V...V..... 07.macFas .............................................................................................................................................................................V...V..... 08.papHam ........................................................................................................................................................L..........................S... 09.cal539 .................N..........I...............................L...V.................................................I.....................................L.............................. 10.cal553 .................N..........I...............................L...V.......................................................................................L.............................. 11.cal561 .................N..........................................L...V...............................I..................H....-...............................L...................V......S... 12.ceb530 .................N..........................................V...F..........V......................................I.....................................L...................V.......... 13.ceb545 .................N..........................................V...V..........V......................-...........I...I.....................................L........M..........V.......... 14.ceb560 .................N..........................................V...F...............................I..................H....................................L...................V......S... 15.ate560 ..........................V.....N.A.........................V...T.................................-...............F.....................................L...................V......S... 16.ate552 ..........................V.....N.A.........................L..............V........................T..I..........F.....................................L...................V......S... 17.tarSyr ,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,.....L...........V..I.........T...........R.......................I..........I.........L...........,,,,,,,,,,,,,,,,,,,,,,,,,,,,......A............ 18.otoGar .....GP...T..Q...TH.....G.........T......................A.........I...........R.......................I..........I.........L...........................L........................V..... 19.otoCra .....GP...T..Q...TH.....G.........T......................A.........I...........R.......................I..........I.........L...........................L........................V..... 20.micMur .....GPH.F..GQ....H.....A.........A......................A.................V...R................I......I..........I.........L...........................L........M........M..T...V..... 21.tupBel ...R.GPHK...GQ..................T.....................I..M.....................R................I.................IF...I................................L.................A............ 22.musMus ...,,,,,,.T.EQTL.H.....HA...........K....................T...L..V..............R.......................I..........I.........L..I...I....................L................T...V...V...I. 23.ratNor ...,,,,,,.T.EQTL.H......A................................T...L.................R.......................I..........I.........L..I...I....................L................T...V...V..... 24.nanEhr .....AP.....GQT.........A.L.............................TT...L..V..I.......V...R.......................I..........I.........L..I.........................................TM..T.A.V..... 25.speTri ...R.DP.....GQ....H...............A...................I..I...I.............V....................I.................FF........L..V......V.................L......................A.T.S... 26.sciCar ,..R.DP.....GQ....H...............A...................I..T...I.............V....................I.................L.........L..V......V.................L........M.................S... 27.dipOrd ,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,...................TI..M..............V...R.......................I..........FN........L.IV........,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 28.cavPor ...R.GPHA.S.VQA..A......A.L.......N......................A..TI.....I.......V...R............................................L..V........................L....................V...V.S... 29.oryCun .T.P.GP.M...GQ.PE.H.....A...................F............A...L.............V...R..................................F.........L..V........................L.................A............ 30.canFam .T.R.GP.....GQ..AGL.E...A.........A..D...................A.................V...R................I......I..........I.........L..V........................L.................A............ 31.phoVit ...T.GP....DGR..PG......A.........A......................T.....................R.......................I......I...I.........L..V.................................................V.S.L. 32.lepWed ,,,,,,,,,,,,,,,,,,,,,,..A.........A...................F..I.....................R.......................I......I...IH........L..V.................................................V.S.L. 33.mirAng ,,,,,,,,,,,,,,,,,,,,,,..A.........A...A..................I.....................R.......................I......I...IH........L..V.................................................V.S.L. 34.eriBar ,,,,,,,,,,,,,,,,,,,,,,..A.........A......................T.....................R.......................I..........IH........L..V.................................................V.S.L. 35.zalCal ,,,,,,,,,,,,,,,,,,,,,,..A.........A......................I.....................R.......................I..........I.........L..V........................L.................A......V.S.L. 36.odoRos ,,,,,,,,,,,,,,,,,,,,,,..A.........A......................T.....................R.......................I..........I.........L..V........................L.................A..V...V.S.L. 37.ursMar ,,,,,,,,,,,,,,,,,,,,,,..A.........A......................A.....................R..................................I.........L..V........................L.................A......V..... 38.enhLut ,,,,,,,,,,,,,,,,,,,,,,..A.........A......................T......F..............R.......................I..........I.........L.............................................A........S... 39.felCat .T.R.GP.....GQ.HAGL....RA.........A...................V..A.............................................I..........I.....................................L.................A............ 40.bosTau ..HA.GP.....GQ..ANF.E...G............D...................A..V..................R................I......I..........M.........L..V..........................................T............ 41.odoVir ..HE.GP.....GQ..ANF.E...G............D...................A..V..................R................I...V..I..........M.........L..V..........................................A........-... 42.turTru .....GP..F..GQ..T.F.....G.V..........D..D...................V..L...I....P.................M.......................M.........L.IV..F.......................................A............ 43.delDel ...T.GP..F..GQ..T.F.....G.V..........D..D...................V..L...I....P.................M.......................M.........L.IV..F.......................................A........-... 44.gloMel .....GP..F..GQ..T.F.....G.V..........D..D...................V..L...I....P.................M.......................M.........L.IV..F.................................................... 45.susScr .....GPR....GQ..A.F.....G.......A....D................I..A..V..................R................I......I..........M.........L..V..........................................A............ 46.equCab ...R.GP.K...GQ..AGF.....A.......N.A..D................V..A.....................R.......................I..........I............V.........................................VA............ 47.myoLuc ...R.GP.....GQL.A.F....LA.........A......................A..V..............V...R.......................L..........I.........L..V........................L........M........A..T...V.S... 48.myoVel ,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,-----.V........................L.................A..T...V.S... 49.pteVam ...S.VP....EGQ..A.F.E...A...I.....V......................A.................V...R.......................I.......A..I.........L..V........................L........M.......T.......V..... 50.pteDas ,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,..V........................L........M.......T.......V..... 51.hapFis ,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,..V........................L............T...T.......V..... 52.sorAra ....,,,,,,,,,,,,RGP.....H....F....A...................V..A......F..............R.......................L....................L...........................L.......SM...............V.S.I. 53.eriEur ...REGP.....VQ...GL.S..LG.T...SG....I..........T...AF..A.A..V...F..............R..............................L...MF......Y.V....C......................L.......SL.....M..L........S... 54.loxAfr .....GPH..T.ARL..AS.....A...V...T.T......................A..L...L..........V...R..................................I....I....L..V........................L.................A..........I. 55.triMan .....GTH..T.GQ...A......A...V...T.T.........H............S..L...M..LL.........VR....................L.............IH...I....L...........................L..........................S.I. 56.echTel ...R.GAH..T.GQL..T..G..RT...V....T...............P.......T......F..........V...R.......................I..........I.....F...L...........................L....................T...V.S.I. 57.proCap ..EP.GTH..T..QLP.AS.....A...V.....A......................A......L..........V...R..................................I....I................................L.................A............ 58.monDom .T.A.DPAGFLA.RRD.DNDET.R..L.V.....N..................N...L..V......I.......V........................G..........I..I....I....L...........................V.........K......M...I...V..... 59.macEug .T.A.DPAGFLAWR-R.EN.ET.RA.L.V.....N.K...............FN...L.........I.......V........................G..L.......I..I....I................................V.........K......M...V...V..... 60.setBra .T.A.DPAGFLAWR-R.EN.ET.RA.L.V.....N.K...............FN...L.........I.......V........................G..M.......I..I....I................................V.........K......M...V...V..... 61.cerCon .T.A.DPAGFLAWQ-E.EN.ET.RA.L.V.....N.K...............FN...L..V......I.......V....................I...G..I.......I..I....I................................V.........K......M...V...V...I. 62.tarRos .T.A.DPAGFLAWR-R.EN.ET.RA.L.V.....N.................FN...L..V......I.......V........................G..I.......I..I....I................................V.........K......M...V...V.-.I. 63.isoObe .T.A.DPAGFLAWR-R.EN.ET.RA.L.V.....N..................N...F..-..............V........................G..I.......I..I....I................................V.........K......M...V...V..... 64.myrFas .T.A.DPAGFLAWRREEN-.ET.RA.L.......N.K................N...F.................V........................G..I.......I..I....I................................V.........K......M...V...V..... 65.ornAna .TPA.NSGVY.A.RRFEDE..T.RT.V.V.....N..D.............A.NV..L.................V........................G..L.......I..IF...I........................S.......I.........K......M...V...V..... 66.tacAcu .T.A.DPAGFLAWR-R.EN.ET.RA.L.V.....N.................FN...L..V......I.......V........................G..I.......I..I....I....L...........................V.........K......M...I...V.-... 67.galGal .-AA.EAA-F.A.RRHEE-..T.RD.V.......N..................N...L......A..........V..W.....................G..........I..IS...I.......V......A....A............F........IK..G...VA..L...L.SCA. 68.taeGut .-AT.DGAVF.A.RRH.D-..T.RD.........N..................N...L......V..........V..A.....................G.............IF...I.......I......A....A............F........IK..G...VA.VL.....SCA. 69.serCar ,,,,,,,,,,,,,,,,,,,,,,,,,,,.......N..................N...L......V..........V..A..............K.S..E.G.............IF...I.......I......A....A............F........IK..G...VA.VL.....SCA. 70.anoCar ..EA.DVAVF.A.RRN.E-D.T.RD.L.......N..................NI............I.......V..A.................I...G..........I..IS...I..............T...SA.....V......V.........K......VA..V...V.S... 71.gecGec .TEA.NVAVF.A.RSR.D-D.T.RG.V.....T.N..................N.V.FF..I.....C.......V..A.................FV..V..LV......F..IF...I....L..I...V..S.................F........IK..S....I..V...V..WG. 72.pheMad .TEA.NVAVF.A.RSR.DDD.T.RG.V....D..N.K...D............NF..F..VI.....T.......V..V.................FV.MV......G...I..IF...I....L..I...L..A.................F........IK..S....L.........WG. 73.utaSta ,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,...A............V.........K......MA..V...V.SC.. 74.ambTig ..HS.NSGAY.A.RRY.D-..T.R..........N..................N..TL......F..........V....................I...G..........I..MF...I....L..I......V...SA....T..A....F........IK..G...AA..I...V.S.G. 75.cynPyr ..YS.NSGAY.A.RRY.D-..T.R..V.V.....N..................N..TL..F...A..........V....................I..I...L.......I..IF...I....L..I......V.........T..A....F........IK..G...AA..I...A...F. 76.xenTro ..SH.NEAVF.A.RRN.D-D.T.R..V.......N..................NIS.L......L..........V..L...............M.I...G..........C..IF..........I.......V...AA....TV.A....F........IK..G...AT..I...V...G. 77.xenLae ..S.LNEAIF.A.RRN.D-D.T.R..V.......N..................N...I......F.........IV..L...............M.I...G..........F..IF...I..........F...T....A....TV.A....F........IK..E...AT..I...V.S.G. 78.neoFor ..EP.D-AV..A.RRHQD-.ET.R.T..V.....N..................N...L......F..C.......M..Y.................I...G..L.......T..IF...I......M...F..AT.........T..A....V........IK..G.W.AG..I...V.S.F. 79.danRer ..EH.GDAIY.A.RKG---DET.REAM.......N.KD...............NVAT...F...V..T.......V..A.................I...G..LF......I..FF...I......IF......V...AA....TV......V.........K....W.SA..I...V...A. 80.cypCar ..E..GDAIF.A.RRG---DET.RETM.V.....N..D...............N.AT...F......T.......V..A.................I......LL......I..IF...I......IF......V...A.....TV......V.........K....W.SA..I...V.S.F. 81.salSal ..ER.GSAAY.A.RQN---Q.T.RE.S..F....N.KD...............N.STL..VI...L.........V..A.....Q...........I..IG..LL......C..FF...I.......F......V...AA.....V......V.......S.K....W.MG..I...V...F. 82.pleAlt .TDE.GNAVF.A.RRN---..T.RE.S.......N.KD...............NISTM.................V..A.....Q...........I...G...L......C..TF...I.......F......T...AA....TV......V.........K....W.TA..V...V.S.A. 83.tetNig ..EE.GK.SF.A.RYH---....RG.A.V.....H..D...............N.ATL..F...VL.........V..A.....................G..LF......C..FF...I......IF...V..V...A.....T.......I.........K....W.TA..V.....PIC. 84.takRub ..EE.GK.SF.A.RYH---..T.RG.A.V.....H..D...............NVAT...FI..VL.........V..A.................I...G...F......C..FF...I.......F......T...AA....T.......V.........K....W.TG..V...V..... 85.gasAcu ..EE.GK.AF.A.RYN---..T.RG.M.V.....N.KD...............N.STL..FI..AL.........V..A.....Q...........I...G...F......C..FF...I.......F...V..V....A....T.......I.........K....W.TA..V.....S... 86.oryLat ..E..GK.VF.A.RQN---..T.RG.A.......H..D...............N.ATL..F...VL.........V..A.............S...I...G...F......C..FF...I.......F...V..T...AA....T.......V.........K....W..G..V...V.S... 87.poeRet ..EE.GK.VF.A.R-H---..T.RGAA.......H.KD...............NVSTL..CI..VL.D.......V..A.................I...G...F......C..FF...I.......F...V..T...AA....T.......I.........K....W.TA..V...A..... 88.lucGoo ..E..EK.AF.A.RYN---.ET.RG.V.......H..D...............NVST...F...VL.........V..A..E..............I...G...F......C..FF...I.......F..FI..T...AA....T.......I.........K....W.TA..V...V.S... 89.ophVen ..EE.GK.SF.A.RYH---....RG.A.A.....N..D.............I.NVATL..FV..VL.........V....................I...G...F......C..FF...I......IF...V..V...AA....T.......I.........K....W.TA..V...V.S... 90.pseMax ..ED.GKPAF.A.RYH---..T.RG.A.M.....H.KD.............I.N.ATL..FV..V..........V..A.................I...G...F......C..FF...I.......F......V...A.....T.......I........IK....W.TG..L.....PI.. 91.calMil ,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,D.............A.N......VG..V..........V..VR..............M.L...G...L...V..A..FF...I....L..F..FV.......A....T..A....V.........K..G.W.AF..I...V.SIG. 92.petMar .TAS.QGAMF.A.RR..D-..T.ME.L.R...E.N.KD..............FN.......I..VL.L.S.....V..V............I....I..IL..IF......C.......I.......F...V..T...A......................IK..G.I.TIL.V...V.P.S. 93.letJap .TAS.HGAVF.A.RRN.D-..T.KD...R...E.N..D.............MFN.......I..VL.L.......V....................I..IL..IF......C...F...I.......F...V..T...A......................IK..G.I..IL.V...V.P.C. 94.geoAus ...S.ERAMF.A.RR..--..T.KGDL.R...E.N..D.............M.N...F...I...L.L.......V..L.................I..IG..IF...V.....IF...I....L..F..F...V....A........F............LK..G.V...L.I...A.S.G. .position 1.....1.........2.........2.........2.........2.........2.........2.........2.........2.........2.........2.........3........3..........3.........3.........3.........3.........3....3 .position 8.....9.........0.........1.........2.........3.........4.........5.........6.........7.........8.........9.........0........1..........2.........3.........4.........5.........6....5 .position 45678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345 .....exon 33333333334444444444444444444444444444444444444444444444444444444555555555555555555555555555555555555555555555555555555555555555555555555555555556666666666666666666666666666666666666 .topology NNNNNNNNNeeeeeeeeeeeeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMMMMMMMMccccccccccccccccccccccNNNNNNNNNNNNNNNNNNNNNNNNeeeeeeeeMMMMMMMMMMMMMMMMMMMMMMMMMccccccccccccccccccccccccccccccccccccccc* ..markers .............P28.diS.........................................................................P-8.....P-15...............P27.....K..................................................... 01.homSap TAPPIFGWSRYWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYLQVWLAIRAVAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQFRNCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA 02.panTro ..............................................I..A..............................................................................................................................--..... 03.gorGor ................................................................................................................................................................................--..... 04.ponPyg ................................................................................................................................................................................--..... 05.nomLeu ..............................................I..A..............................................................................................................................--..... 06.macMul .................................................T........................................F..Y.V.....T............A..........Y..........I.......................................--..... 07.macFas .................................................T.....................................M..IF.Y.V.....T............A..........Y..........I.......................................--..... 08.papHam .................................................T........................................F.......................A..........Y..........I.......................................--..... 09.cal539 .........................................I....L...........................................IA.Y.V.....T............A..........Y..........I.......................................--..... 10.cal553 .........................................I...FL...........................................IA.Y.V.....T............A..........Y..........I.......................................--..... 11.cal561 .........................................I...FL..G........................................IV.Y.V.....T............A..........Y..........I.......................................--..... 12.ceb530 .........................................I....L...........................................IVT..V..................A..........Y..........I.......................................--..... 13.ceb545 .........................................I....L...........................................I.T..V.....T............A..........Y..........I.......................................--..... 14.ceb560 .........................................I...FL..G........................................IM.Y.V.....T............A..........Y..........I.......................................--..... 15.ate560 .............................D...K...........FL..G........................................IM.Y.V.....T............A..........Y..................................................--..... 16.ate552 .............................D................L...........................................IM...I.....T........K...A..........Y..................................................--..... 17.tarSyr .................................................T.........................................F.Y.L.....T........H...A....V.....Y..........I..........................V............--..... 18.otoGar .....................................M.......VI.....M........................................Y.V.............SH...A....V.....Y..........I.......................S.....T.........--..... 19.otoCra .....................................M.......VI.....M........................................Y.V.............SH...A....V.....Y..........I.......................S.....T.........--..... 20.micMur ......................................M.......I..............................................Y.V..R..T........H...A..........Y..........I.............................T.........--..... 21.tupBel ..............................................I............................................C.Y.V.....T........H...A....L.....Y..........I.......................S.......R..A....--..... 22.musMus .............Y.............T.........M........F............................................F.Y.L.....T......T.H...A....V.S..SY..........I..............H........S.....T.........--..... 23.ratNor .............Y.............T.........M........F............................................F.Y.L.....T......T.H...A....V.S..SY..........I.......................S.....T.........--..... 24.nanEhr ....V........Y.............T.........V...I....I.....L...........T...................H......F.Y.L.....T..V...T.H...A....V.....Y..........I.........Q.............S.....T....A....--..... 25.speTri .............Y.............N.........M........I.....I........................................Y.L......S.....T.....A....V..V..Y..................................T.......R..A....--..... 26.sciCar .............Y.............T....M....M........I.....I......................................F.Y.L.....T......T.....A....V.....Y..........I,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 27.dipOrd ,,,,,,,,,,...Y.............T.........M........I.....................L......................F.Y.V............T.H...A....V....SY..........I.......................S.....T....A....--..... 28.cavPor .............Y.............T.........M...................H...................................Y.L............T.....S....V.....Y..........I.....................E.S.....T.R..A....--..... 29.oryCun .............Y.............T.........M........I...V.........M..P...........................F.Y.L.....T......T.H...S....V..I.SY..........I.....................E.S.......R..A....--..... 30.canFam .........................................T....I...V.I........................................Y.L.....T........H...A....V.....Y..........I.......................S.......R..A....--..... 31.phoVit .........................................T....I..GV.I......................................F...V.....T........H...A....V.....Y..........I.......................S..........A....--..... 32.lepWed .........................................T....I..GV.I.....................,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 33.mirAng .........................................T....I..GV.I..................D,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 34.eriBar .........................................T....I..GV.I.....................,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 35.zalCal .........................................T....I...V.I....................,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 36.odoRos .........................................T....I...V.I.......................,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 37.ursMar .........................................T....I...V.I.....................,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 38.enhLut .........................................I....I.....I.......................,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 39.felCat .........................................I....I...V....................................M..IF.Y.V.....T........H...A....V.....Y..........I.............M.................R..A....--..... 40.bosTau .........................................I...FI...V.I..................................M..IF.Y.L.....T........H...A....V.....Y..........I.......................S.....V....A....--..... 41.odoVir .........................................I...FI...V....................................M..IF...L........,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 42.turTru ......................................T..I...FI...V.........................R..........M..IF.Y.L.....T........H...A....V....SY..........I.......................S.....V....A....--..... 43.delDel ......................................T..I...FI...VT........................R.V........M..IF.Y.L.....T........H...A....V....SY..........I.......................S.....V....A....--..... 44.gloMel ......................................T..I...FI...V.........................R..........M..IF.Y.L.....T........H...A....V....SY..........I.......................S.....V....A....--..T.. 45.susScr .........................................I...FI..GV....................................M..IF.Y.L.....T........H...A....V.....Y..........I.......................S.....V....A....--..... 46.equCab .........................................I....I...V....................................M...F...L.....T........H...A....V.....Y..........I.......................S.....V....A....--..... 47.myoLuc .........................................T....I...V.......................................I..Y.L.....T........H...A....V.....Y..........I...................R...S.....T.R..A....--..... 48.myoVel .........................................T....I...V.......................................I..Y.L.....T........H...A....V.....Y..........I........,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 49.pteVam ..............................................I...V.LF.......................................Y.L.....T........H...A....V.....Y..........I.......................S.....S....A....--..... 50.pteDas ..............................................I...V.LF.......................................Y.L.....T........H...A....V.....Y..........I........,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 51.hapFis ..............................................I...V..F.......................................Y.L.....T........H...A....V.....Y..........I........,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 52.sorAra ...........................D.............I....I..GV.LF.................................M.....Y.V.....T........H...A....V.....Y..........I.............................V..S.AT...--..... 53.eriEur .............................D.......M........I......F...H...................................Y.V.....T.S.S....H...T....L.T..SY...................,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 54.loxAfr .........................................T....I..........H................................I..Y.L.....T......T.H...A..........Y..........I.......................S..........A....--..... 55.triMan .....................................M...I....I..G.....-.H.............................L..I..Y.......T........H...A....V.....Y..........I..................................A....--..... 56.echTel .................................R.......I....I........................................M..IF.Y.L.....TI.......H...S..........Y..................................S.....T.R..A....--..... 57.proCap ................................A........I....I..........H.............................M..IF.Y.V.....T........H...A..........Y..........I.......................S.....T..M.A..A.--..... 58.monDom ....L........................D...........A....F.....L...V..........................S......I..Y.......TL...........S....T.S...Y..........I..........T...............V..T.R.......--..A.. 59.macEug ....L......................N.D...........S....L...V.F...I.......S..................S......I...........I...........A....T.S...Y..........I..........T...............V..T.R.......--..A.. 60.setBra ....L......................N.D...........S....L...V.L...I..........................S......I..Y........I...........A....T.S...Y..........I..........T...............V..T.R.......--..PA. 61.cerCon .S..L......................N.D..I........S....L.....L...I..........................S......I..Y.......T............A....T.S...Y..........I..........T...............V..T.R.......--..A.. 62.tarRos .S..L......................N.D..I........S....L.....L...V...R......................S......I..Y.......TL...........A....T.S...Y..........I..........T...............V..T.R.......--..A.. 63.isoObe .............................D...........T....L.....L...V..............D...........S......IR.Y.......TL...........A....T.S...Y..........I..........T...............V.GT.R.......--.APA 64.myrFas .............................D...........S....L...V.L...I..........................S......I..Y........I...........A....T.S...Y..........I..........T...............V..T.R.......--..A.. 65.ornAna .............................D...........S....L....................................S......I..Y.......TI...........A....A.....Y..........I.............M...............T.R.......--..... 66.tacAcu .S..L........................D...........A....F.....L...I..........................S......I..Y.......TL...........A....T.S...Y..........I..........T...............V..T.........--..A.. 67.galGal .............................D.......V.......FF..A..I......S.......A...............S......IV.Y.......T............A....A.....Y..........I..........................V.-T.R.......NS..... 68.taeGut ............................TD....-..V.......FF..AV.IF.............A...............S......I..Y.......TI...........A....T................I..........................V.-T.R.......NS..... 69.serCar ...........................TTD.......V.......FF..AV.IF.............A...............S......I..Y.......TI..S......-.A....T................I........,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,, 70.anoCar ....V.......................DD...L.......I...FI..AV.L..............A...............S......II.Y.......TV...........A....A.....Y..........I.............M...............T.R.......NS..... 71.gecGec S...........................VEL.C..F.LT..I...FL..F..IV......M......A............R..S......IV...I......S.VS........A....A.....Y........................M............A.TT.R.......NS..A.. 72.pheMad S..........................NNEL.C....LA...S..FF...V.I.......M......A......................L....V.....TI.VG....H...A....A.....Y.......W.....I....................A.DV.TT.R.......NS..... 73.utaSta ............................DD...........I...FI..AV.I.......M......A............R..S......II.Y.......TI...........A....A.....Y..........I.............................T.R.......NS..... 74.ambTig C............................D.......M...I....L.....II..I...W...Q..M............R..S......II.YI......T............A....A.S..............I.............Y............M....R.......NS..... 75.cynPyr C..........................N.D.....F..T..S....L.....I...V...W...Q..M............R..S......IM.YI......T..V.........S....A.S...Y..........I.............Y...............S.R.......NS..... 76.xenTro C............................D.......L...I....I..A......MH...T..Q..Q............R..S......II.YI......T.......F....N....A..M..Y..........I.............Y............V..T.R.......NS..... 77.xenLae C...M.....F..................D.......L...I....I..A..I....H..WT..Q..Q............R..S......IV.YI......T.......FS...S....A.....Y..........I.............Y.M..........V..T.R.......NS..... 78.neoFor C.M.L.....F................EDKY.TR.F..A..I....I..GV.I...I...W...T..................S......IF.Y.......T.M...G..Y...A....A.....Y..........I.............Y..L............T.........NS..... 79.danRer C...........................ED.......V...I....I..A..I...IA.Y...H...Q...D...........S......IF.Y.......T............A....A..M..Y.....................V..M............V.-T.......-----.A.. 80.cypCar C.........F.................ED...........I....I..A..I...IA.........Q...D...........S......IF.Y.......T............A....A..M..Y..........I..........V..M............V.-T.......-----.A.. 81.salSal C........................G.NED...K....T..I...FF..FV.IF..IF.........A...D...........S......II.Y.V.....TC...........A....A..I..Y.....................T..M.....AE...T.V.-T.......-----.A.. 82.pleAlt C...........................DD...K.......I...FL..A..I...IA.........Q...D...........S......I..YIV.....TV...........A....A.....Y.....................V..M............V.-T.......-----.A.. 83.tetNig C...........................ED...........I....I..A.......A..M......M............R..S......I..Y.V.....T............A....A..M..Y..........I..........V..MK....E......V.-T.......-----.A.. 84.takRub C...........................ED...........I....I..A..I....A......S..M...............S......IV.Y.V.....T............A....A..M..Y.....................V..MK....E......V.-T.......-----.A.. 85.gasAcu C...........................ED...........I...LI..A..I....A.........M............RD.S......IV.YIV.....TT...........A....A..M..Y.....................S..M.....E......V.-T.......-----.A.. 86.oryLat C...V.......................DD...........I....I..A..I....A.........M............R..S......IV.Y.V.....T............A....A..M..Y.....................T..M.....Q......V.-T.......-----.A.. 87.poeRet C...........................DD...L.......I....I..A..I....A.........M............R..S......II.Y.V.....T.....P......A....A..M..Y.....................T..M.....Q....F.V.-T.......-----.A.. 88.lucGoo C...........................ED.......V...I....I..A..I....A.........M............R.GS......I..Y.V.....T............A....A..M..Y...................S.T..M.....Q......V.-T.......-----.A.. 89.ophVen C...........................ED...........I....L..AV.I....A.........M............R..S......IV.Y.V.....T............A....A..M..Y..........I..........T..M.....Q......V.-T.......-----.A.. 90.pseMax C...........................ED...............FL..A..I....A..W..HS..L.............D.S......I..Y.V.....T............A....A..M..Y..........I..........T..M.....E......V.-T.......-----.A.. 91.calMil CL..V.....,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,.....S.......A...L......C..M.S.L....A....V.SI.SY....S.....I.............................T...D.....NS..... 92.petMar CSL.........................TD.......V...I...FL.....I..........HS..Q......T.....RD.S......I..YV......T............S...IA.....Y...G......I..........................V..S.R.......NS..... 93.letJap CSL..........................D.......V.......FL...V.I..........HS..Q......T.....RD.S......I..YI......T....Y.......A....T.....Y........................M............V....R.......NS.I... 94.geoAus C...........................TD.......V...I...FI..AL.II.........HT..Q......T.....RD.S......IF.YI......T............A....A.....Y..........I.............M............V..SAR.......NS.....
=== The marsupial insert in exon 1
For example, this novel insertion in the first exon of LWS is a pretty good one for uniting the Didelphimorphia to the exclusion of the other marsupials: LWS_homSap MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS LWS_gorGor MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS LWS_ponPyg MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS LWS_panTro MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS LWS_nomLeu MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS LWS_papHam MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS LWS_macMul MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS LWS_macFas MAQQWSLQRLAGRHPQDSYE---DSTQSSIFTYTNSNS LWS_cal539 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFIYTNSNS LWS_cal553 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFIYTNSNS LWS_ceb530 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFTYTNSNS LWS_cal561 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFTYTNSNS LWS_ceb560 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFTYTNSNS LWS_ceb545 MAQQWSLQRLAGRHPQDNYE---DSTQSSIFTYTNSNS LWS_ate560 MAQQWSLQRLAGRHPQDSYE---DSTQSSVFTYTNNNA LWS_ate552 MAQQWSLQRLAGRHPQDSYE---DSTQSSVFTYTNNNA LWS_otoGar MAQQWGPQRLTGRQPQDTHE---DSTQGSIFTYTNSNT LWS_otoCra MAQQWGPQRLTGRQPQDTHE---DSTQGSIFTYTNSNT LWS_micMur MAQQWGPHRFAGGQPQDSHE---DSTQASIFTYTNSNA LWS_tupBel MAQRWGPHKLAGGQPQDSYE---DSTQSSIFTYTNTNS LWS_musMus MAQ------LTGEQTLDHYE---DSTHASIFTYTNSNS LWS_ratNor MAQ------LTGEQTLDHYE---DSTQASIFTYTNSNS LWS_nanEhr MAQQWAPQRLAGGQTQDSYE---DSTQASLFTYTNSNS LWS_speTri MAQRWDPQRLAGGQPQDSHE---DSTQSSIFTYTNSNA LWS_oryCun MTQPWGPQMLAGGQPPESHE---DSTQASIFTYTNSNS LWS_cavPor MAQRWGPHALSGVQAQDAYE---DSTQASLFTYTNSNN LWS_canFam MTQRWGPQRLAGGQPQAGLE---ESTQASIFTYTNSNA LWS_felCat MTQRWGPQRLAGGQPHAGLE---DSTRASIFTYTNSNA LWS_bosTau MAHAWGPQRLAGGQPQANFE---ESTQGSIFTYTNSNS LWS_susScr MAQQWGPRRLAGGQPQASFE---DSTQGSIFTYTNANS LWS_equCab MAQRWGPQKLAGGQPQAGFE---DSTQASIFTYTNNNA LWS_phoVit MAQTWGPQRLADGRPQPGYE---DSTQASIFTYTNSNA LWS_myoLuc MAQRWGPQRLAGGQLQASFE---DSTLASIFTYTNSNA LWS_pteVam MAQSWVPQRLAEGQPQASFE---ESTQASIFIYTNSNV LWS_turTru MAQQWGPQRFAGGQPQTSFE---DSTQGSVFTYTNSNS LWS_gloMel MAQQWGPQRFAGGQPQTSFE---DSTQGSVFTYTNSNS LWS_delDel MAQTWGPQRFAGGQPQTSFE---DSTQGSVFTYTNSNS LWS_loxAfr MAQQWGPHRLTGARLQDASE---DSTQASIFVYTNTNT LWS_proCap MAEPWGTHRLTGRQLPDASE---DSTQASIFVYTNSNA LWS_echTel MAQRWGAHRLTGGQLQDTYE---GSTRTSIFVYTNSTS LWS_triMan MAQQWGTHRLTGGQPQDAYE---DSTQASIFVYTNTNT LWS_monDom MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNN Didelphimorphia LWS_didAur MTQAWDPVGFLARRRDENEDDHDDTTRASLFVYTNSNN Didelphimorphia LWS_tarRos MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodontia LWS_macEug MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodontia LWS_smiCra MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Dasyuromorphia LWS_myrFas MTQAWDPAGFLAWRREENE----ETTRASLFTYTNSNN Dasyuromorphia LWS_setBra MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodonti LWS_cerCon MTQAWDPAGFLAWQEDENE----ETTRASLFVYTNSNN Diprotodontia LWS_isoObe MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Peramelemorphia LWS_ornAna MTPAWNSGVYAARRRFEDEE---DTTRTSVFVYTNSNN LWS_tacAcu MTQAWDPAGFLAWRRDENEE---TTRASLFVYTNSNNT LWS_galGal MAAWEAA--FAARRRHEEE----DTTRDSVFTYTNSNN LWS_taeGut MATWDGAV-FAARRRHDDE----DTTRDSIFTYTNSNN LWS_colLiv M---DG---FAAARRRHEDE---DTTRDSVFTYTNSNN LWS_anoCar MAEAWDVAVFAARRRNDED----DTTRDSLFTYTNSNN LWS_gecGec MTEAWNVAVFAARRSRDDD----DTTRGSVFTYTNTNN LWS_pheMad MTEAWNVAVFAARRSRDDDD---DTTRGSVFTYTDSNN LWS_cynPyr MAYSWNSGAYAARRRYDDE----DTTRSSVFVYTNSNN LWS_ambTig MAHSWNSGAYAARRRYDDE----DTTRSSIFTYTNSNN LWS_xenTro MASHWNEAVFAARRRNDDD----DTTRSSVFTYTNSNN LWS_xenLae MASQLNEAIFAARRRNDDD----DTTRSSVFTYTNSNN LWS_neoFor MAEPWD-AVLAARRRHQDE----ETTRSTIFVYTNSNN LWS_danRer MAEHWGDAIYAARRKGD------ETTREAMFTYTNSNN LWS_cypCar MAEQWGDAIFAARRRGD------ETTRETMFVYTNSNN LWS_pleAlt MTDEWGNAVFAARRRNE------DTTRESSFTYTNSNN LWS_tetNig MAEEWGKQSFAARRYHE------DSTRGSAFVYTNSNH LWS_takRub MAEEWGKQSFAARRYHE------DTTRGSAFVYTNSNH LWS_ophVen MAEEWGKQSFAARRYHE------DSTRGSAFAYTNSNN LWS_oryLat MAEQWGKQVFAARRQNE------DTTRGSAFTYTNSNH LWS_lucGoo MAEQWEKQAFAARRYNE------ETTRGSVFTYTNSNH LWS_gasAcu MAEEWGKQAFAARRYNE------DTTRGSMFVYTNSNN LWS_pseMax MAEDWGKPAFAARRYHE------DTTRGSAFMYTNSNH LWS_poeRet MAEEWGKQVFAARR-HE------DTTRGAAFTYTNSNH LWS_salSal MAERWGSAAYAARRQNQ------DTTRESSFTFTNSNN LWS1_calMi MTQSWELVAPAARRGFKYD----EPTHSGIFVYTNSNQ LWS2_calMi MAEPRGSVAFAARR-WNDH----EGTTVGEFTYTNSNS LWS_sciCar MAQRWDPQRLAGGQPQDSHE---DSTQSSIFTYTNSNA LWS_eriEur MAQREGPQRLAGVQPQDGLE---SSTLGSTFTYSGSNS LWS_petMar MTASWQGAMFAARRRQDDE----DTTMESLFRYTNENN LWS_letJap MTASWHGAVFAARRRNDDE----DTTKDSIFRYTNENN LWS_geoAus MAQSWERAMFAARRRQDE-----DTTKGDLFRYTNENN
Indels in the cytoplasmic tail
LWS fixed four indels during vertebrate history: a 2 residue loss in mammals, a 1 residue loss in birds but not lizards, and a 1 and 5 residue loss in teleost fish. The event in bird, if pursued in more species, might be helpful in defining subclades, though note the site experienced a homoplasic event in teleost fish. Otherwise, the cytoplasmic tail of LWS has been remarkably constant -- its key features and almost every residue past FR were already fixed by the time of lamprey divergence.
LWS_homSap KSATIYNPVIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_gorGor KSATIYNPVIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_ponPyg KSATIYNPVIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_panTro KSATIYNPVIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_nomLeu KSATIYNPVIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_papHam KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_macMul KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_macFas KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_cal539 KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_cal553 KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_ceb530 KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_cal561 KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_ceb560 KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_ceb545 KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_ate560 KSATIYNPVIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_ate552 KSATIYNPVIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEVSSVS--SVSPA LWS_otoGar KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSTSKTEVSSVS--SVSPA LWS_otoCra KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSTSKTEVSSVS--SVSPA LWS_micMur KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSTSKTEVSSVS--SVSPA LWS_tupBel KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSASRTEASSVS--SVSPA LWS_canFam KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSASRTEASSVS--SVSPA LWS_felCat KSATIYNPIIYVFMNRQFR NCIMQLFGKKVDDGSELSSASRTEASSVS--SVSPA LWS_bosTau KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSVSKTEASSVS--SVSPA LWS_susScr KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSVSKTEASSVS--SVSPA LWS_equCab KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSVSKTEASSVS--SVSPA LWS_phoVit KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSASKTEASSVS--SVSPA LWS_myoLuc KSATIYNPIIYVFMNRQFR NCILQLFGKRVDDSSELSSTSRTEASSVS--SVSPA LWS_pteVam KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSSSKTEASSVS--SVSPA LWS_turTru KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSVSKTEASSVS--SVSPA LWS_gloMel KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSVSKTEASSVS--SVTPA LWS_delDel KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSVSKTEASSVS--SVSPA LWS_loxAFR KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSASKTEASSVS--SVSPA LWS_proCap KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSTSKMEASSAS--SVSPA LWS_echTel KSATIYNPVIYVFMNRQFR NCILQLFGKKVDDSSELSSTSRTEASSVS--SVSPA LWS_speTri KSATIYNPVIYVFMNRQFR NCILQLFGKKVDDTSELSSASRTEASSVS--SVSPA LWS_oryCun KSATIYNPIIYVFMNRQFR NCILQLFGKKVEDSSELSSASRTEASSVS--SVSPA LWS_cavPor KSATIYNPIIYVFMNRQFR NCILQLFGKKVEDSSELSSTSRTEASSVS--SVSPA LWS_triMan KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSASKTEASSVS--SVSPA LWS_musMus KSATIYNPIIYVFMNRQFR NCILHLFGKKVDDSSELSSTSKTEVSSVS--SVSPA LWS_ratNor KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSTSKTEVSSVS--SVSPA LWS_nanEhr KSATIYNPIIYVFMNRQFQ NCILQLFGKKVDDSSELSSTSKTEASSVS--SVSPA LWS_sorAra KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSVSKSEATSVS--SVSPA LWS_monDom KSATIYNPIIYVFMNRQFR TCILQLFGKKVDDGSEVSSTSRTEVSSVS--SVAPA LWS_myrFas KSATIYNPIIYVFMNRQFR TCILQLFGKKVDDGSEVSSTSRTEVSSVS--SVAPA LWS_macEug KSATIYNPIIYVFMNRQFR TCILQLFGKKVDDGSEVSSTSRTEVSSVS--SVAPA LWS_smiCra KSATIYNPIIYVFMNRQFR TCILQLFGKKVDDGSEVSSTSRTEVSSVS--SVAPA LWS_setBra KSATIYNPIIYVFMNRQFR TCILQLFGKKVDDGSEVSSTSRTEVSSVS--SV-PA LWS_cerCon KSATIYNPIIYVFMNRQFR TCILQLFGKKVDDGSEVSSTSRTEVSSVS--SVAPA LWS_tarRos KSATIYNPIIYVFMNRQFR TCILQLFGKKVDDGSEVSSTSRTEVSSVS--SVAPA LWS_tacAcu KSATIYNPIIYVFMNRQFR TCILQLFGKKVDDGSEVSSTSKTEVSSVS--SVAPA LWS_isoObe KSATIYNPIIYVFMNRQFR TCILQLFGKKVDDGSEVSGTSRTEVSSVS--S-APA LWS_tarSyr KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSEVSSASKTEVSSVS--SVSPA LWS_dipOrd KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDSSELSSTSKTEASSVS--SVSPA LWS_ornAna KSATIYNPIIYVFMNRQFR NCIMQLFGKKVDDGSELSSTSRTEVSSVS--SVSPA LWS_galGal KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSEVS-TSRTEVSSVSNSSVSPA LWS_taeGut KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSEVS-TSRTEVSSVSNSSVSPA LWS_colLiv KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSEVS-TSRTEVSSVSSSSVSPA LWS_anoCar KSATIYNPIIYVFMNRQFR NCIMQLFGKKVDDGSELSSTSRTEVSSVSNSSVSPA LWS_gecGec KSATIYNPVIYVFMNRQFR NCIMQLFGKKVDDGSEASTTSRTEVSSVSNSSVAPA LWS_pheMad KSATIWNPVIYIFMNRQFR NCILQLFGKKVDDASDVSTTSRTEVSSVSNSSVSPA LWS_utaSta KSATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSTSRTEVSSVSNSSVSPA LWS_ambTig KSATIYNPIIYVFMNRQFR NCIYQLFGKKVDDGSEMSSASRTEVSSVSNSSVSPA LWS_cynPyr KSATIYNPIIYVFMNRQFR NCIYQLFGKKVDDGSELSSSSRTEVSSVSNSSVSPA LWS_xenTro KSATIYNPIIYVFMNRQFR NCIYQLFGKKVDDGSEVSSTSRTEVSSVSNSSVSPA LWS_xenLae KSATIYNPIIYVFMNRQFR NCIYQMFGKKVDDGSEVSSTSRTEVSSVSNSSVSPA LWS_neoFor KSATIYNPIIYVFMNRQFR NCIYQLLGKKVDDGSELSSTSKTEVSSVSNSSVSPA LWS_danRer KSATIYNPVIYVFMNRQFR VCIMQLFGKKVDDGSEVS-TSKTEVSS-----VAPA LWS_cypCar KSATIYNPIIYVFMNRQFR VCIMQLFGKKVDDGSEVS-TSKTEVSS-----VAPA LWS_pleAlt KSATIYNPVIYVFMNRQFR VCIMQLFGKKVDDGSEVS-TSKTEVSS-----VAPA LWS_tetNig KSATIYNPIIYVFMNRQFR VCIMKLFGKEVDDGSEVS-TSKTEVSS-----VAPA LWS_takRub KSATIYNPVIYVFMNRQFR VCIMKLFGKEVDDGSEVS-TSKTEVSS-----VAPA LWS_ophVen KSATIYNPIIYVFMNRQFR TCIMQLFGKQVDDGSEVS-TSKTEVSS-----VAPA LWS_oryLat KSATIYNPVIYVFMNRQFR TCIMQLFGKQVDDGSEVS-TSKTEVSS-----VAPA LWS_lucGoo KSATIYNPVIYVFMNRQSR TCIMQLFGKQVDDGSEVS-TSKTEVSS-----VAPA LWS_gasAcu KSATIYNPVIYVFMNRQFR SCIMQLFGKEVDDGSEVS-TSKTEVSS-----VAPA LWS_pseMax KSATIYNPIIYVFMNRQFR TCIMQLFGKEVDDGSEVS-TSKTEVSS-----VAPA LWS_poeRet KSATIYNPVIYVFMNRQFR TCIMQLFGKQVDDGFEVS-TSKTEVSS-----VAPA LWS_salSal KSATIYNPVIYVFMNRQFR TCIMQLFGKAEDDGTEVS-TSKTEVSS-----VAPA LWS_calMil KSSTIYNPIIYVFMNRQFR NCILQLFGKKVDDGSELSSTSKTDVSSVSNSSVSPA LWS_petMar KGATIYNPIIYVFMNRQFR NCILQLFGKKVDDGSEVSSSSRTEVSSVSNSSVSPA LWS_letJap KSATIYNPVIYVFMNRQFR NCIMQLFGKKVDDGSEVSSASRTEVSSVSNSSISPA LWS_geoAus KSATIYNPIIYVFMNRQFR NCIMQLFGKKVDDGSEVSSSARTEVSSVSNSSVSPA
Curated LWS reference sequences from 95 species
>LWS_homSap Homo sapiens (human) -IRAK1 -MECP2 -TEX28 +TKTL1 P530 full 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_panTro Pan troglodytes (chimp) odd full 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 gPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVSGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIIGIAFSWIWSAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCIIPLAIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_gorGor Gorilla gorilla (gorilla) full 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_ponPyg Pongo pygmaeus (orang_sumatran) full 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVTASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVSGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_nomLeu Nomascus leucogenys (gibbon) full 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVTASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVSGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIIGIAFSWIWSAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCIIPLAIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_macMul Macaca mulatta (rhesus) full 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVSGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLTIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMFLAYCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_macFas Macaca fascicularis (macaque) full 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLTIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVMVMIFAYCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* >LWS_papHam Papio hamadras (baboon) full 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFSWIWSAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLTIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMFLAFCFCWGPYAFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_cal539 Callithrix jacchus (marmoset) P539 full 0 MAQQWSLQRLAGRHPQDNYEDSTQSSIFIYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMLFVVVASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQIYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCILPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMIAAYCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_cal553 Callithrix jacchus (marmoset) P553 full 0 MAQQWSLQRLAGRHPQDNYEDSTQSSIFIYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMLFVVVASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCFLPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMIAAYCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_cal561 Callithrix jacchus (marmoset) P561 full 0 MAQQWSLQRLAGRHPQDNYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMLFVVVASVFTNGLVLAATMKFKKLRHPLNWILVNLAIADLAETVIASTISVVNQVHGYFVGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGVAFSWIWSAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCFLPLGIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_ceb530 Cebus capucinus P530 full 0 MAQQWSLQRLAGRHPQDNYEDSTQSSIFTYTNSNSTR 1 2 gPFEGPNYHIAPRWVYHLTSVWMVFVVFASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQIYGYFVLGHPMCVLEGYTVSLC 1 2 gITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGVAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCILPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMIVTFCVCWGPYAFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_ceb545 Cebus capucinus P545 full 0 MAQQWSLQRLAGRHPQDNYEDSTQSSIFTYTNSNSTR 1 2 gPFEGPNYHIAPRWVYHLTSVWMVFVVVASVFTNGLVLVATMKFKKLRHPLNWILVNLAVALAETVIASTISIVNQIYGYFVLGHPMCVLEGYTVSLC 1 2 gITGLWSLAIISWERWLVVCKPFGNMRFDAKLAIVGVAFSWIWAAVWTAPPIFGWSr 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCILPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMILTFCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_ceb560 Cebus capucinus P560 full 0 MAQQWSLQRLAGRHPQDNYEDSTQSSIFTYTNSNSTR 1 2 gPFEGPNYHIAPRWVYHLTSVWMVFVVFASVFTNGLVLAATMKFKKLRHPLNWILVNLAIADLAETVIASTISVVNQVHGYFVLGHPMCVLEGYTVSLC 1 2 gITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGVAFSWIWSAVWTAPPIFGWSr 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCFLPLGIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMIMAYCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_ate560 Ateles geoffroyi P560 full 0 MAQQWSLQRLAGRHPQDSYEDSTQSSVFTYTNNNATR 1 2 gPFEGPNYHIAPRWVYHLTSVWMVFVVTASVFTNGLVLAATMKFKKLRHPLNWILVNLAVALAETVIASTISVVNQFYGYFVLGHPMCVLEGYTVSLC 1 2 gITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGVAFSWIWSAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVKSYMIVLMVTCCFLPLGIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMIMAYCVCWGPYTFFACFAAANPGYAFHPLMAALPAYFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_ate552 Ateles geoffroyi P552 full 0 MAQQWSLQRLAGRHPQDSYEDSTQSSVFTYTNNNATR 1 2 gPFEGPNYHIAPRWVYHLTSVWMLFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLTETIIASTISVVNQFYGYFVLGHPMCVLEGYTVSLC 1 2 gITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGVAFSWIWSAVWTAPPIFGWSr 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMVTCCILPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMIMAFCICWGPYTFFACFAAAKPGYAFHPLMAALPAYFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_tarSyr Tarsius syrichta (tarsier) frag 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 1 2 GPFEGLNYHIAPRWVYHVTSIWMIFVVIASTFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVLEGYTVSLC 1 2 zzzzzzzzzzzzzzzzzzzzzzzzzzzzDAKLAIAGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLTIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAkSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSEVSSASKTEVSSVSSVSPA* 0 >LWS_otoGar Otolemur garnettii (bushbaby) full 0 MAQQWGPQRLTGRQPQDTHEDSTQGSIFTYTNSNTTR 1 2 GPFEGPNYHIAPRWVYHLTSAWMIFVVIASIFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMMVLMVTCCVIPLSIIMLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCVCWGPYAFFACFAASHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA* 0 >LWS_otoCra Otolemur crassicaudatus full 0 MAQQWGPQRLTGRQPQDTHEDSTQGSIFTYTNSNTTR 1 2 GPFEGPNYHIAPRWVYHLTSAWMIFVVIASIFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMMVLMVTCCVIPLSIIMLCYLQVWLAIRA 0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCVCWGPYAFFACFAASHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA* 0 >LWS_micMur Microcebus murinus (mouse_lemur) full 0 MAQQWGPHRFAGGQPQDSHEDSTQASIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHLTSAWMIFVVIASVFTNGLVLVATMRFKKLRHPLNWILVNLAIADLAETIIASTISVVNQIYGYFVLGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNMRFDAKLAIMGITFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIMLMVTCCIIPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCVCWRPYTFFACFAAAHPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRq 0 0 FRNCILQLFGKKVDDGSELSSTSKTEVSSVSSVSPA* 0 >LWS_tupBel Tupaia belangeri (tree_shrew)full 0 MAQRWGPHKLAGGQPQDSYEDSTQSSIFTYTNTNSTR 1 2 GPFEGPNYHIAPRWVYHITSMWMIFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAIADLAETVIASTISVVNQIFGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCIIPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVCAYCVCWGPYTFFACFAAAHPGYAFHPLLAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSASRTEASSVSSVSPA* 0 >LWS_musMus Mus musculus (mouse) full 0 MAQzzzzzzLTGEQTLDHYEDSTHASIFTYTNSNSTK 1 2 GPFEGPNYHIAPRWVYHLTSTWMILVVVASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYIVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLATVGIVFSWVWAAIWTAPPIFGWSR 2 1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCIFPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATAHPGYAFHPLVASLPSYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILHLFGKKVDDSSELSSTSKTEVSSVSSVSPA* 0 >LWS_ratNor Rattus norvegicus (rat) full 0 MAQzzzzzzLTGEQTLDHYEDSTQASIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSTWMILVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYIVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLATVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCIFPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATAHPGYAFHPLVASLPSYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA* 0 >LWS_nanEhr Nannospalax ehrenbergi (mole_rat) full 0 MAQQWAPQRLAGGQTQDSYEDSTQASLFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTTTWMILVVVASIFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLATMGITFAWVWAAVWTAPPVFGWSR 2 1 YWPYGLKTSCGPDVFSGTSYPGVQSYMVVLMITCCIIPLSIILLCYLQVWLAIRT 2 0 VAKQQKESESTQKAEKEVTHMVVVMVFAYCLCWGPYTFFVCFATAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FQNCILQLFGKKVDDSSELSSTSKTEASSVSSVSPA* 0 >LWS_speTri Spermophilus tridecemlineatus (squirrel) full 0 MAQRWDPQRLAGGQPQDSHEDSTQSSIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHITSIWMIIVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAIADLAETVIASTISVVNQFFGYFVLGHPLCVVEGYTVSVC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFAWTWSAVWTAPPIFGWSR 2 1 YWPYGLKTSCGPDVFSGNSYPGVQSYMMVLMVTCCIIPLSIIILCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCLCWGPYASFACFATANPGYAFHPLVAAVPAYFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDTSELSSASRTEASSVSSVSPA* 0 >LWS_sciCar Sciurus carolinensis (squirrel) frag 0 zAQRWDPQRLAGGQPQDSHEDSTQSSIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHITSTWMIIVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAIADLAETVIASTISVVNQLYGYFVLGHPLCVVEGYTVSVC 1 2 GITGLWSLAIISWERWLVVCKPFGNMRFDAKLAIVGIAFSWIWSAVWTAPPIFGWSR 2 1 YWPYGLKTSCGPDVFSGTSYPGMQSYMMVLMVTCCIIPLSIIILCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATANPGYAFHPLVAALPAYFAKSATIYNPIzzzzzzzz 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_dipOrd Dipodomys ordii (pika) frag 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 1 2 GPFEGPNYHIAPRWVYHLTTIWMMFVVIASVFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQFNGYFVLGHPLCIVEGYTVSLC 1 2 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 2 1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCIIPLSIIVLCYLQVWLAIRA 0 0 VAKLQKESESTQKAEKEVTRMVVVMVFAYCVCWGPYAFFACFATAHPGYAFHPLVAALPSYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSTSKTEASSVSSVSPA* 0 >LWS_cavPor Cavia porcellus (guinea_pig) full 0 MAQRWGPHALSGVQAQDAYEDSTQASLFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYHLTSAWMTIVVIASIFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIVFSWVWSAVWTAPPIFGWSR 2 1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCITPLSIIVLCYLHVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCLCWGPYAFFACFATANPGYSFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVEDSSELSSTSRTEASSVSSVSPA* 0 >LWS_oryCun Oryctolagus cuniculus (rabbit) full 0 MTQPWGPQMLAGGQPPESHEDSTQASIFTYTNSNSTR 1 2 GPFEGPNFHIAPRWVYHLTSAWMILVVIASVFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETVIASTISVVNQFYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCIIPLSVIVLCYLQVWMAIPA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATAHPGYSFHPLVAAIPSYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVEDSSELSSASRTEASSVSSVSPA* 0 >LWS_canFam Canis familiaris (dog) full 0 MTQRWGPQRLAGGQPQAGLEESTQASIFTYTNSNATR 1 2 DPFEGPNYHIAPRWVYHLTSAWMIFVVIASVFTNGLVLVATMRFKKLRHPLNWILVNLAIADLAETIIASTISVVNQIYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLSVIILCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSASRTEASSVSSVSPA* 0 >LWS_phoVit Phoca vitulina (seal) full 0 MAQTWGPQRLADGRPQPGYEDSTQASIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHLTSTWMIFVVIASVFTNGLVLaATMRFKKLRHPLNWILVNLAVADLAETIIASTISIVNQIYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWVWSALWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLGVIILCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVFAFCVCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSASKTEASSVSSVSPA* 0 >LWS_lepWed Leptonychotes weddellii (seal) frag 0 zzzzzzzzzzzzzzzzzzzzzzTQASIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHFTSIWMIFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISIVNQIHGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWVWSALWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLGVIILCYLQVWLAIRA 0 0 VAKQQKESEzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_mirAng Mirounga angustirostris (elephant seal) frag 0 zzzzzzzzzzzzzzzzzzzzzzTQASIFTYTNSNATR 1 2 GAFEGPNYHIAPRWVYHLTSIWMIFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISIVNQIHGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWVWSALWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLGVIILCYLQVWLAIRA 0 0 VAKQQKDzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_eriBar Erignathus barbatus (seal) frag 0 zzzzzzzzzzzzzzzzzzzzzzTQASIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHLTSTWMIFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIHGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWVWSALWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLGVIILCYLQVWLAIRA 0 0 VAKQQKESEzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_zalCal Zalophus californianus (sea lion) frag 0 zzzzzzzzzzzzzzzzzzzzzzTQASIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHLTSIWMIFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIAFSWVWSALWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLSVIILCYLQVWLAIRA 0 0 VAKQQKESzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_odoRos Odobenus rosmarus (walrus) frag 0 zzzzzzzzzzzzzzzzzzzzzztqASIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHLTSTWMIFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIVFSWVWSALWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLSVIILCYLQVWLAIRA 0 0 VAKQQKESESTzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_ursMar Ursus maritimus (polar bear) frag 0 zzzzzzzzzzzzzzzzzzzzzzTQASIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHLTSAWMIFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETVIASTISVVNQIYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIAFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLSVIILCYLQVWLAIRA 0 0 VAKQQKESEzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_enhLut Enhydra lutris (sea otter) frag 0 zzzzzzzzzzzzzzzzzzzzzzTQASIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHLTSTWMIFVVFASVFTNGLVLAATMRFKkLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIAGIAFSWIWSAVWTAPPIFGWSR 2 1 YWPHGLKTScGPDVFSGSSYPGVQSYMIVLMITCCIIPLSIIILCYLQVWLAIRA 0 0 VAKQQKESESTzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_felCat Felis catus (cat) full 0 MTQRWGPQRLAGGQPHAGLEDSTRASIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHVTSAWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCIIPLSVIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVMVMIFAYCVCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSELSSASRTEASSVSSVSPA* 0 >LWS_bosTau Bos taurus (cow) full 0 MAHAWGPQRLAGGQPQANFEESTQGSIFTYTNSNSTR 1 2 DPFEGPNYHIAPRWVYHLTSAWMVFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAIADLAETIIASTISVVNQMYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAITGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCFIPLSVIILCYLQVWLAIRA 0 VAKQQKESESTQKAEKEVTRMVMVMIFAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA* 0 >LWS_odoVir Odocoileus virginianus (deer) frag 0 MAHEWGPQRLAGGQPQANFEESTQGSIFTYTNSNSTR 1 2 DPFEGPNYHIAPRWVYHLTSAWMVFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAIADLVETIIASTISVVNQMYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIAGIAFSWIWAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCFIPLSVIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVMVMIFAFCLCWGPYAFFzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_turTru Tursiops truncatus (dolphin) full 0 MAQQWGPQRFAGGQPQTSFEDSTQGSVFTYTNSNSTR 1 2 DPFDGPNYHIAPRWVYHLTSVWMVFVLIASIFTNGPVLAATMKFKKLRHPLNWMLVNLAVADLAETVIASTISVVNQMYGYFVLGHPLCIVEGFTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIAGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMITLMITCCFIPLSVIVLCYLQVWLAIRA 0 0 VAKQQKESESTRKAEKEVTRMVMVMIFAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPSYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA* 0 >LWS_delDel Delphinus delphis (dolphin) full 0 MAQTWGPQRFAGGQPQTSFEDSTQGSVFTYTNSNSTR 1 2 DPFDGPNYHIAPRWVYHLTSVWMVFVLIASIFTNGPVLAATMKFKKLRHPLNWMLVNLAVADLAETVIASTISVVNQMYGYFVLGHPLCIVEGFTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIAGIAFSWIWAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMITLMITCCFIPLSVTVLCYLQVWLAIRA 0 0 VAKQQKESESTRKVEKEVTRMVMVMIFAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPSYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA* 0 >LWS_gloMel Globicephala melas (pilot_whale) full 0 MAQQWGPQRFAGGQPQTSFEDSTQGSVFTYTNSNSTR 1 2 DPFDGPNYHIAPRWVYHLTSVWMVFVLIASIFTNGPVLAATMKFKKLRHPLNWMLVNLAVADLAETVIASTISVVNQMYGYFVLGHPLCIVEGFTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMITLMITCCFIPLSVIVLCYLQVWLAIRA 0 0 VAKQQKESESTRKAEKEVTRMVMVMIFAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPSYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVTPA* 0 >LWS_susScr Sus scrofa (pig) full 0 MAQQWGPRRLAGGQPQASFEDSTQGSIFTYTNANSTR 1 2 DPFEGPNYHIAPRWVYHITSAWMVFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAIADLAETIIASTISVVNQMYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIAGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCFIPLGVIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVMVMIFAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA* 0 >LWS_equCab Equus caballus (horse) full 0 MAQRWGPQKLAGGQPQAGFEDSTQASIFTYTNNNATR 1 2 DPFEGPNYHIAPRWVYHVTSAWMIFVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPMCVVEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAVAGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMITCCIIPLSVIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVMVMVFAFCLCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA* 0 >LWS_myoLuc Myotis lucifugus (brown bat) full 0 MAQRWGPQRLAGGQLQASFEDSTLASIFTYTNSNATR 1 2 GPFEGPNYHIAPRWVYHLTSAWMVFVVIASVFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETLIASTISVVNQIYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNMRFDAKLAIAGITFSWVWSAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLSVIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMILAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKRVDDSSELSSTSRTEASSVSSVSPA* 0 >LWS_myoVel Myotis velifer (bat) AM263194 mRNA frag 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 1 2 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzxxxxxVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGITFSWVWSAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLSVIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMILAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_pteVam Pteropus vampyrus (flying fox) full 0 MAQSWVPQRLAEGQPQASFEESTQASIFIYTNSNVTR 1 2 GPFEGPNYHIAPRWVYHLTSAWMIFVVIASVFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETIIASTISVANQIYGYFVLGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNMRFDAKLATVGIAFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCIIPLSVILFCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSSSKTEASSVSSVSPA* 0 >LWS_pteDas Pteropus dasymallus formosus (bat) AM263195 mRNA frag 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 1 2 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNMRFDAKLATVGIAFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCIIPLSVILFCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_hapFis Haplonycteris fischeri (fruit_bat) mRNA frag 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 1 2 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDTKLATVGIAFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCIIPLSVIVFCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCLCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_sorAra Sorex araneus (shrew)full 0 MAQQzzzzzzzzzzzzRGPEDSTQHSIFTFTNSNATR 1 2 GPFEGPNYHIAPRWVYHVTSAWMIFVVFASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETLIASTISVVNQVYGYFVLGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGSMRFDAKLAIVGIAFSWVWSAIWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGDSYPGVQSYMIVLMITCCIIPLGVILFCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVMVMVLAYCVCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSVSKSEATSVSSVSPA* 0 >LWS_eriEur Erinaceus europaeus (hedgehog)frag 0 MAQREGPQRLAGVQPQDGLESSTLGSTFTYSGSNSTI 1 2 GPFEGPNYHITPRWAFHLASAWMVFVVFASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETVIASTISLVNQMFGYFVLGYPVCVLECYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGSLRFDAKMAILGIAFSWIWSAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMMVLMVTCCIIPLSIIVFCYLHVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCVCWGPYTFSASFAAAHPGYTFHPLLATLPSYFAKSATIYNPVIYVFMNRQ 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_loxAfr Loxodonta africana (elephant) full 0 MAQQWGPHRLTGARLQDASEDSTQASIFVYTNTNTTR 1 2 GPFEGPNYHIAPRWVYHLTSAWMLFVVLASVFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETVIASTISVVNQIYGYFILGHPLCVVEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIAFSWIWAAIWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMTTCCIIPLSIIVLCYLHVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMILAYCLCWGPYTFFACFATAHPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSASKTEASSVSSVSPA* 0 >LWS_triMan Trichechus manatus (manatee) full 0 MAQQWGTHRLTGGQPQDAYEDSTQASIFVYTNTNTTR 1 2 GPFEGPNHHIAPRWVYHLTSSWMLFVVMASLLTNGLVLAATVRFKKLRHPLNWILVNLAVADLLETVIASTISVVNQIHGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFSWIWSAIWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMMVLMITCCIIPLGIIVLCLHVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVLVMILAYCFCWGPYTFFACFAAAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEASSVSSVSPA* 0 >LWS_echTel Echinops telfairi (tenrec) full 0 MAQRWGAHRLTGGQLQDTYEGSTRTSIFVYTNSTSTR 1 2 GPFEGPNYHIAPPWVYHLTSTWMIFVVFASVFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVFGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGITFSWVWSAIWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVRSYMIVLMITCCIIPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVMVMIFAYCLCWGPYTIFACFAAAHPGYSFHPLMAALPAYFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSTSRTEASSVSSVSPA* 0 >LWS_proCap Procavia capensis (hyrax) full 0 MAEPWGTHRLTGRQLPDASEDSTQASIFVYTNSNATR 1 2 GPFEGPNYHIAPRWVYHLTSAWMIFVVLASVFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETVIASTISVVNQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGAQSYMIVLMITCCIIPLSIIVLCYLHVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVMVMIFAYCVCWGPYTFFACFAAAHPGYAFHPLMAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDSSELSSTSKMEASSASSVSPA* 0 >LWS_monDom Monodelphis domesticus (opossum) Didelphimorphia) VNED insert 0 MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYVQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYSFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSKTEGSSVSSVAPA* 0 >LWS_didAur Didelphis aurita (big-eared opossum) Didelphimorphia ENED insert 0 MTQAWDPVGFLARRRDENEDDHDDTTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDLGVQSYMIVLMATCCIFPLSIILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA* 0 >LWS_tarRos Tarsipes rostratus (honey possum) Diprotodontia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAIWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGIQSYMIVLMSTCCILPLSIILLCYVQVWRAIRA 2 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_macEug Macropus eugenii (wallaby) Diprotodontia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGVQSYMIVLMSTCCILPLSVIFLCYIQVWLAIRS 2 0 VAKQQKESESTQKAEKEVSRMVVVMILAFCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_setBra Setonix brachyurus (quokka) Diprotodontia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETMIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGVQSYMIVLMSTCCILPLSVILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVPA* 0 >LWS_cerCon Cercartetus concinnus (pygmy possum) Diprotodontia 0 MTQAWDPAGFLAWQEDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAIADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAIWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGIQSYMIVLMSTCCILPLSIILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTFFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* >LWS_smiCra Sminthopsis crassicaudata (dunnart) Dasyuromorphia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIILCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVMVMILAFCFCWGPYALFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_myrFas Myrmecobius fasciatus (numbat) Dasyuromorphia 0 MTQAWDPAGFLAWRREENEETTRASLFTYTNSNNTK 1 2 GPFEGPNYHIAPRWVYNLTSFWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSVILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_isoObe Isoodon obesulus (bandicoot) Peramelemorphia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSFWMFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMTTCCILPLSIILLCYVQVWLAIRA 0 0 VAKQQKDSESTQKAEKEVSRMVVVMIRAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSGTSRTEVSSVSSAPA* 0 >LWS_ornAna Ornithorhynchus anatinus (platypus) full 0 MTPAWNSGVYAARRRFEDEEDTTRTSVFVYTNSNNTR 1 2 DPFEGPNYHIAPRWAYNVTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIFGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLSIISWERWIVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSSVSPA* 0 >LWS_tacAcu Tachyglossus aculeatus (echidna) full 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAVWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA* 0 >LWS_galGal Gallus gallus (chicken) missing genome full 0 MAAWEAAFAARRRHEEEDTTRDSVFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVAASVFTNGLVLVATWKFKKLRHPLNWILVNLAVADLGETVIASTISVINQISGYFILGHPMCVVEGYTVSAC 1 2 GITALWSLAIISWERWFVVCKPFGNIKFDGKLAVAGILFSWLWSCAWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFFPLAIIILCYLQVSLAIRA 0 0 VAAQQKESESTQKAEKEVSRMVVVMIVAYCFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSEVSTSRTEVSSVSNSSVSPA* 0 >LWS_taeGut Taeniopygia guttata (finch) GV excised full 0 MAT WDGAVFAARRRHDDEDTTRDSIFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVVASVFTNGLVLVATAKFKKLRHPLNWILVNLAVADLGETVIASTISVVNQIFGYFILGHPMCVIEGYTVSAC 1 2 GITALWSLAIISWERWFVVCKPFGNIKFDGKLAVAGVLFSWIWSCAWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSTDPGVQYMVVLMVTCCFFPLAVIIFCYLQVWLAIRA 0 0 VAAQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFACFAAANPGYAFHPLTAALPAFFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSEVSTSRTEVSSVSNSSVSPA* 0 >LWS_colLiv Columba livia (pigeon) P559 full 0 MDGFAAARRRHEDEDTTRDSVFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVVASVFTNGLVLVATWRFKKLRHPLNWILVNLAVADLGETVIASTISVVNQISGYFVLGHPMCVLEGYTVSAC 1 2 GITALWSLAIISWERWFVVCKPFGNIKFDGKLALAGILFSWVWSCAWTAPPVFGWSR 2 0 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFFPLAIIVLCYLQVWLAIRA 0 0 VAAQQKESESTQKAEKEVSRMVVVMIVAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSEVSTSRTEVSSVSSSSVSPA* 0 >LWS_serCar Serinus canaria (canary) frag 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVVASVFTNGLVLVATAKFKKLRHPLNWILVKLSVAELGETVIASTISVVNQIFGYFILGHPMCVIEGYTVSAC 1 2 GITALWSLAIISWERWFVVCKPFGNIKFDGKLAVAGVLFSWIWSCAWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGTTDPGVQSYMVVLMVTCCFFPLAVIIFCYLQVWLAIRA 0 0 VAAQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFASFAAANPYAFHPLTAALPAFFAKSATIYNPIIYVFMNRQ 0 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz* 0 >LWS_anoCar Anolis carolinensis (lizard) GTVT excised full 0 MA EAWDVAVFAARRRNDEDDTTRDSLFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNITSVWMIFVVIASIFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVIASTISVINQISGYFILGHPMCVLEGYTVSTC 1 2 GISALWSLAVISWERWVVVCKPFGNVKFDAKLAVAGIVFSWVWSAVWTAPPVFGWSR 2 1 YWPHGLKTSCGPDVFSGSDDPGVLSYMIVLMITCCFIPLAVILLCYLQVWLAIRA 0 0 VAAQQKESESTQKAEKEVSRMVVVMIIAYCFCWGPYTVFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSNSSVSPA* 0 >LWS_gecGec Gecko gecko (gecko) P521 full 0 MTEAWNVAVFAARRSRDDDDTTRGSVFTYTNTNNTR 1 2 GPFEGPNYHIAPRWVYNLVSFFMIIVVIASCFTNGLVLVATAKFKKLRHPLNWILVNLAFVDLVETLVASTISVFNQIFGYFILGHPLCVIEGYVVSSC 1 2 GITGLWSLAIISWERWFVVCKPFGNIKFDSKLAIIGIVFSWVWAWGWSAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSVELGCQSFMLTLMITCCFLPLFIIIVCYLQVWMAIRA 0 0 VAAQQKESESTQKAEREVSRMVVVMIVAFCICWGPYASFVSFAAANPGYAFHPLAAALPAYFAKSATIYNPVIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSEASTTSRTEVSSVSNSSVAPA* 0 >LWS_pheMad Phelsuma madagascariensis (gecko) full 0 MTEAWNVAVFAARRSRDDDDDTTRGSVFTYTDSNNTK 1 2 GPFDGPNYHIAPRWVYNFTSFWMVIVVIASTFTNGLVLVATVKFKKLRHPLNWILVNLAFVDMVETVIASGISVINQIFGYFILGHPLCVIEGYLVSAC 1 2 GITGLWSLAIISWERWFVVCKPFGNIKFDSKLAILGIAFSWIWAWGWSAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGNNELGCQSYMLALMVSCCFFPLSVIILCYLQVWMAIRA 0 0 VAAQQKESESTQKAEKEVTRMVVVMLLAFCVCWGPYTIFVGFAAAHPGYAFHPLAAALPAYFAKSATIWNPVIYIFMNRQ 0 0 FRNCILQLFGKKVDDASDVSTTSRTEVSSVSNSSVSPA* 0 >LWS_utaSta Uta stansburiana (lizard) frag 0 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz1 2 zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz 1 2 GITALWSLAIISWERWVVVCKPFGNVKFDAKLAMAGIVFSWVWSCVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSDDPGVQSYMIVLMITCCFIPLAVIILCYLQVWMAIRA 0 0 VAAQQKESESTQKAEREVSRMVVVMIIAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSTSRTEVSSVSNSSVSPA* 0 >LWS_ambTig Ambystoma tigrinum (salamander) full 0 MAHSWNSGAYAARRRYDDEDTTRSSIFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTTLWMIFVVFASVFTNGLVLVATMKFKKLRHPLNWILVNLAIADLGETVIASTISVINQMFGYFILGHPLCVIEGYTVSVC 1 2 GISALWSLTIIAWERWFVVCKPFGNIKFDGKLAAAGIIFSWVWSAGWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMMVLMITCCILPLSIIIICYIQVWWAIRQ 0 0 VAMQQKESESTQKAEREVSRMVVVMIIAYIFCWGPYTFFACFAAANPGYAFHPLAASLPAFFAKSATIYNPIIYVFMNRQ 0 0 FRNCIYQLFGKKVDDGSEMSSASRTEVSSVSNSSVSPA* 0 >LWS_cynPyr Cynops pyrrhogaster (newt) full 0 MAYSWNSGAYAARRRYDDEDTTRSSVFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTTLWMFFVVAASVFTNGLVLVATMKFKKLRHPLNWILVNLAIADIAETLIASTISVINQIFGYFILGHPLCVIEGYTVSVC 1 2 GITGLWSLTIIAWERWFVVCKPFGNIKFDGKLAAAGIIFSWAWAAFWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGVQSFMITLMSTCCILPLSIIILCYVQVWWAIRQ 0 0 VAMQQKESESTQKAEREVSRMVVVMIMAYIFCWGPYTFFVCFAAANPGYSFHPLAASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIYQLFGKKVDDGSELSSSSRTEVSSVSNSSVSPA* 0 >LWS_xenTro Xenopus tropicalis (frog) full 0 MASHWNEAVFAARRRNDDDDTTRSSVFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNISSLWMIFVVLASVFTNGLVLVATLKFKKLRHPLNWILVNMAIADLGETVIASTISVCNQIFGYFVLGHPMCILEGYTVSVC 1 2 GIAALWSLTVIAWERWFVVCKPFGNIKFDGKLAATGIIFSWVWAAGWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMLVLMITCCIIPLAIIVLCYMHVWLTIRQ 0 0 VAQQQKESESTQKAEREVSRMVVVMIIAYIFCWGPYTFFACFAAFNPGYNFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIYQLFGKKVDDGSEVSSTSRTEVSSVSNSSVSPA* 0 >LWS_xenLae Xenopus laevis (frog) full 0 MASQLNEAIFAARRRNDDDDTTRSSVFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSIWMIFVVFASVFTNGLVIVATLKFKKLRHPLNWILVNMAIADLGETVIASTISVFNQIFGYFILGHPMCVLEGFTVSTC 1 2 GITALWSLTVIAWERWFVVCKPFGNIKFDEKLAATGIIFSWVWSAGWCAPPMFGWSR 2 1 FWPHGLKTSCGPDVFSGSSDPGVQSYMLVLMITCCIIPLAIIILCYLHVWWTIRQ 0 0 VAQQQKESESTQKAEREVSRMVVVMIVAYIFCWGPYTFFACFAAFSPGYSFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIYQMFGKKVDDGSEVSSTSRTEVSSVSNSSVSPA* 0 >LWS_neoFor Neoceratodus forsteri (lungfish) full 0 MAEPWDAVLAARRRHQDEETTRSTIFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVFASCFTNGLVLMATYKFKKLRHPLNWILVNLAIADLGETLIASTISVTNQIFGYFILGHPMCMLEGFTVATC 1 2 GITGLWSLTIIAWERWVVVCKPFGNIKFDGKWAAGGIIFSWVWSAFWCAMPLFGWSR 2 1 FWPHGLKTSCGPDVFSGEDKYGTRSFMIALMITCCIIPLGVIILCYIQVWWAIRT 0 0 VAKQQKESESTQKAEKEVSRMVVVMIFAYCFCWGPYTFMACFGAAYPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIYQLLGKKVDDGSELSSTSKTEVSSVSNSSVSPA* 0 >LWS_danRer Danio rerio (zebrafish) full 0 MAEHWGDAIYAARRKGDETTREAMFTYTNSNNTK 1 2 DPFEGPNYHIAPRWVYNVATVWMFFVVVASTFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETLFASTISVINQFFGYFILGHPMCIFEGYTVSVC 1 2 GIAALWSLTVISWERWVVVCKPFGNVKFDAKWASAGIIFSWVWAAAWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMVVLMITCCIIPLAIIILCYIAVYLAIHA 2 0 VAQQQKDSESTQKAEKEVSRMVVVMIFAYCFCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 0 FRVCIMQLFGKKVDDGSEVSTSKTEVSSxxxxxVAPA* 0 >LWS_cypCar Cyprinus carpio (carp) full 0 MAEQWGDAIFAARRRGDETTRETMFVYTNSNNTR 1 2 DPFEGPNYHIAPRWVYNLATVWMFFVVIASTFTNGLVLVATAKFKKLRHPLNWILVNLAIADLAETLLASTISVINQIFGYFILGHPMCIFEGYTVSVC 1 2 GIAGLWSLTVISWERWVVVCKPFGNVKFDAKWASAGIIFSWVWSAFWCAPPIFGWSR 2 1 FWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCIIPLAIIILCYIAVWLAIRA 0 0 VAQQQKDSESTQKAEKEVSRMVVVMIFAYCFCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0 0 FRVCIMQLFGKKVDDGSEVSTSKTEVSSxxxxxVAPA* 0 >LWS_salSal Salmo salar (salmon) full 0MAERWGSAAYAARRQNQDTTRESSFTFTNSNNTK 1 2 DPFEGPNYHIAPRWVYNLSTLWMVIVVILSVFTNGLVLVATAKFKKLQHPLNWILVNLAIADIGETLLASTISVCNQFFGYFILGHPMCVFEGYTVSVC 1 2 GIAALWSLAVISWERWVVVCKPFGSVKFDAKWAMGGIIFSWVWAAFWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFGGNEDPGVKSYMITLMITCCFFPLFVIIFCYIFVWLAIRA 0 0 VAAQQKDSESTQKAEKEVSRMVVVMIIAYCVCWGPYTCFACFAAANPGYAFHPLAAAIPAYFAKSATIYNPVIYVFMNRQ 0 0 FRTCIMQLFGKAEDDGTEVSTSKTEVSSxxxxxVAPA* 0 >LWS_pleAlt Plecoglossus altivelis (ayu) full 0 MTDEWGNAVFAARRRNEDTTRESSFTYTNSNNTK 1 2 DPFEGPNYHIAPRWVYNISTMWMIFVVIASVFTNGLVLVATAKFKKLQHPLNWILVNLAIADLGETVLASTISVCNQTFGYFILGHPMCVFEGYTVSTC 1 2 GIAALWSLTVISWERWVVVCKPFGNVKFDAKWATAGIVFSWVWSAAWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSDDPGVKSYMIVLMITCCFLPLAIIILCYIAVWLAIRA 0 0 VAQQQKDSESTQKAEKEVSRMVVVMILAYIVCWGPYTVFACFAAANPGYAFHPLAAALPAYFAKSATIYNPVIYVFMNRQ 0 0 FRVCIMQLFGKKVDDGSEVSTSKTEVSSxxxxxVAPA* 0 >LWS_tetNig Tetraodon nigroviridis (pufferfish) full 0 MAEEWGKQSFAARRYHEDSTRGSAFVYTNSNHTR 1 2 DPFEGPNYHIAPRWVYNLATLWMFFVVVLSVFTNGLVLVATAKFKKLRHPLNWILVNLAVADLGETLFASTISVCNQFFGYFILGHPMCIFEGYVVSVC 1 2 GIAGLWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWIWPICWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCIIPLAIIVLCYLAVWMAIRA 2 0 VAMQQKESESTQKAEREVSRMVVVMILAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0 0 FRVCIMKLFGKEVDDGSEVSTSKTEVSSxxxxxVAPA* 0 >LWS_takRub Takifugu rubripes (pufferfish) full 0 MAEEWGKQSFAARRYHEDTTRGSAFVYTNSNHTR 1 2 DPFEGPNYHIAPRWVYNVATVWMFIVVVLSVFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYTVSTC 1 2 GIAALWSLTIISWERWVVVCKPFGNVKFDAKWATGGIVFSWVWAAVWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCIIPLAIIILCYLAVWLAIRS 0 0 VAMQQKESESTQKAEKEVSRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 0 FRVCIMKLFGKEVDDGSEVSTSKTEVSSxxxxxVAPA* 0 >LWS_gasAcu Gasterosteus aculeatus (stickleback) full 0 MAEEWGKQAFAARRYNEDTTRGSMFVYTNSNNTK 1 2 DPFEGPNYHIAPRWVYNLSTLWMFIVVALSVFTNGLVLVATAKFKKLQHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYVVSVC 1 2 GITALWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWIWSAVWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCLIPLAIIILCYLAVWLAIRA 0 0 VAMQQKESESTQKAERDVSRMVVVMIVAYIVCWGPYTTFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 0 FRSCIMQLFGKEVDDGSEVSTsKTEVSSxxxxxVAPA* 0 >LWS_oryLat Oryzias latipes (medaka) full 0 MAEQWGKQVFAARRQNEDTTRGSAFTYTNSNHTR 1 2 DPFEGPNYHIAPRWVYNLATLWMFFVVVLSVFTNGLVLVATAKFKKLRHPLNWILSNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYVVSTC 1 2 GIAALWSLTIISWERWVVVCKPFGNVKFDAKWAIGGIVFSWVWSAVWCAPPVFGWSR 2 1 YWPHGLKTSCGPDVFSGSDDPGVQSYMIVLMITCCIIPLAIIILCYLAVWLAIRA 0 0 VAMQQKESESTQKAEREVSRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 0 FRTCIMQLFGKQVDDGSEVSTSKTEVSSxxxxxVAPA* 0 >LWS_poeRet Poecilia reticulata (guppy) full 0 MAEEWGKQVFAARRHEDTTRGAAFTYTNSNHTK 1 2 DPFEGPNYHIAPRWVYNVSTLWMCIVVVLSDFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYVVSTC 1 2 GIAALWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWAWAAVWCAPPIFGWS 2 1 RYWPHGLKTSCGPDVFSGSDDPGVLSYMIVLMITCCIIPLAIIILCYLAVWLAIRA 0 0 VAMQQKESESTQKAEREVSRMVVVMIIAYCVCWGPYTFFACFPAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 0 FRTCIMQLFGKQVDDGFEVSTSKTEVSSxxxxxVAPA* 0 >LWS_lucGoo Lucania goodei (killifish) full 0 MAEQWEKQAFAARRYNEETTRGSVFTYTNSNHTR 1 2 DPFEGPNYHIAPRWVYNVSTVWMFFVVVLSVFTNGLVLVATAKFEKLRHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGFIVSTC 1 2 GIAALWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWVWSAVWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMVVLMITCCIIPLAIIILCYLAVWLAIRAVA 0 0 MQQKESESTQKAEREGSRMVVVMILAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 0 SRTCIMQLFGKQVDDGSEVSTSKTEVSSxxxxxVAPA* 0 >LWS_ophVen Ophthalmotilapia ventralis (cichlid) full 0 MAEEWGKQSFAARRYHEDSTRGSAFAYTNSNNTR 1 2 DPFEGPNYHIAPRWIYNVATLWMFVVVVLSVFTNGLVLVATMKFKKLRHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCIFEGYVVSVC 1 2 GIAALWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWVWSAVWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCILPLAVIILCYLAVWLAIRA 0 0 VAMQQKESESTQKAEREVSRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCIMQLFGKQVDDGSEVSTSKTEVSSxxxxxVAPA* 0 .>LWS_pseMax Psetta maxima (turbot) full 0 MAEDWGKPAFAARRYHEDTTRGSAFMYTNSNHTK 1 2 DPFEGPNYHIAPRWIYNLATLWMFVVVVASVFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYTVSVC 1 2 GIAGLWSLTIISWERWIVVCKPFGNIKFDAKWATGGILFSWIWPIVWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMVTCCFLPLAIIILCYLAVWWAIHS 0 0 VALQQKESESTQKAEKDVSRMVVVMILAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCIMQLFGKEVDDGSEVSTSKTEVSSxxxxxVAPA* 0 >LWS1_calMil Callorhinchus milii (elephantfish) EF565165 0 MTQSWELVAPAARRGFKYDEPTHSGIFVYTNSNQTR 1 2 GPFEGPNYHIAPRWAYNLTSVWMVGVVVASVFTNGLVLVATVRFKKLRHPLNWILVNMALADLGETVLASTVSVANQFFGYFILGHPLCVFEGFVVSLC 1 2 GITALWSLTIIAWERWVVVCKPFGNMKFDSKMAVAGIVFSWVWSAGWCLPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGNEDPGVQSYMVALTLSCAVLPLLIIILCYFQVWWAIRA 0 0 VALQQKESESTQKAEKEVSRMVVVMVAAFCLCWGPYACFAMFSALNPGYAFHPLVASIPSYFAKSSTIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSTSKTDVSSVSNSSVSPA* 0 >LWS2_calMil Callorhinchus milii (elephantfish) EF565166 0 MAEPRGSVAFAARRWNDHEGTTVGEFTYTNSNSTR 1 2 DPFEGPNYHIAPRWTYNLTSLWMVVVVILSVFTNGLVLVATWKFKKLRHPLNWILVNLAIADLGETLFASTISICNQVFGYFILGHPMCVFEGFTVSAC 1 2 GITALWSLTIIAWERWVVVCKPFGNVKFDGKWAAFGIIFSWVWSIGWCLPPVFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVKSYMVTLVITCAALPLTIIIVCLYQVWLAIRA 0 0 VAMQQKESESTQKAEKEVSRMVVVMIIAFCFCWGPYTSFAVFSALNPGYSFHPLMAALPAYFAKSSTIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSTSKTDVSSVSNSSVSPA* 0 >LWS_petMar Petromyzon marinus (lamprey) full 0 MTASWQGAMFAARRRQDDEDTTMESLFRYTNENNTK 1 2 DPFEGPNYHIAPRWVFNLTSVWMIIVVVLSLFSNGLVLVATVKFKKLRHPLNWIIVNLAIADILETIFASTISVCNQVYGYFILGHPMCVFEGYVVSTC 1 2 GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIATILIVFSWVWPASWCSLPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFLPLSIIILCYLQVWLAIHS 0 0 VAQQQKESETTQKAERDVSRMVVVMILAYVFCWGPYTFFACFAAANPGYSFHPIAAALPAYFAKGATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSEVSSSSRTEVSSVSNSSVSPA* 0 >LWS_letJap Lethenteron japonicum (lamprey) full 0 MTASWHGAVFAARRRNDDEDTTKDSIFRYTNENNTR 1 2 DPFEGPNYHIAPRWMFNLTSVWMIIVVVLSLFTNGLVLVATMKFKKLRHPLNWILVNLAIADILETIFASTISVCNQVFGYFILGHPMCVFEGYVVSTC 1 2 GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIAIILIVFSWVWPACWCSLPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFLPLSVIILCYLQVWLAIHS 0 0 VAQQQKESETTQKAERDVSRMVVVMILAYIFCWGPYTFFACYAAANPGYAFHPLTAALPAYFAKSATIYNPVIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSEVSSASRTEVSSVSNSSISPA* >LWS_geoAus Geotria australis (lamprey) full 0 MAQSWERAMFAARRRQDEDTTKGDLFRYTNENNTR 1 2 DPFEGPNYHIAPRWMYNLTSFWMIIVVILSLFTNGLVLVATLKFKKLRHPLNWILVNLAIADIGETIFASTVSVVNQIFGYFILGHPLCVFEGFTVSVC 1 2 GITALWSLAIISFERWMVVCKPFGNLKFDGKVAIVLIIFSWAWSAGWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFIPLALIIICYLQVWLAIHT 0 0 VAQQQKESETTQKAERDVSRMVVVMIFAYIFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSEVSSSARTEVSSVSNSSVSPA* 0
See also: Curated Sequences | Encephalopsins | Melanopsins | Neuropsin | Peropsin | RGR phyloSNPs | Update Blog